Research
/Security News
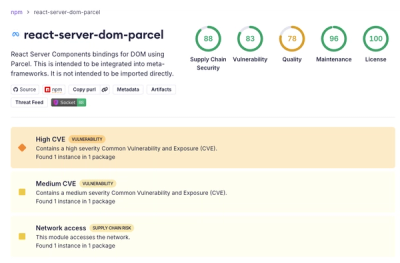
Malicious NuGet Package Typosquats Popular .NET Tracing Library to Steal Wallet Passwords
Impostor NuGet package Tracer.Fody.NLog typosquats Tracer.Fody and its author, using homoglyph tricks, and exfiltrates Stratis wallet JSON/passwords to a Russian IP address.