@jbrowse/img
Static exports of JBrowse 2 rendering.
Prerequisites
You don't need to have JBrowse 2 installed to use this tool. The tool can
generate images using files on your hard drive or from remote files. So, all you
need to run this tool is
Screenshot
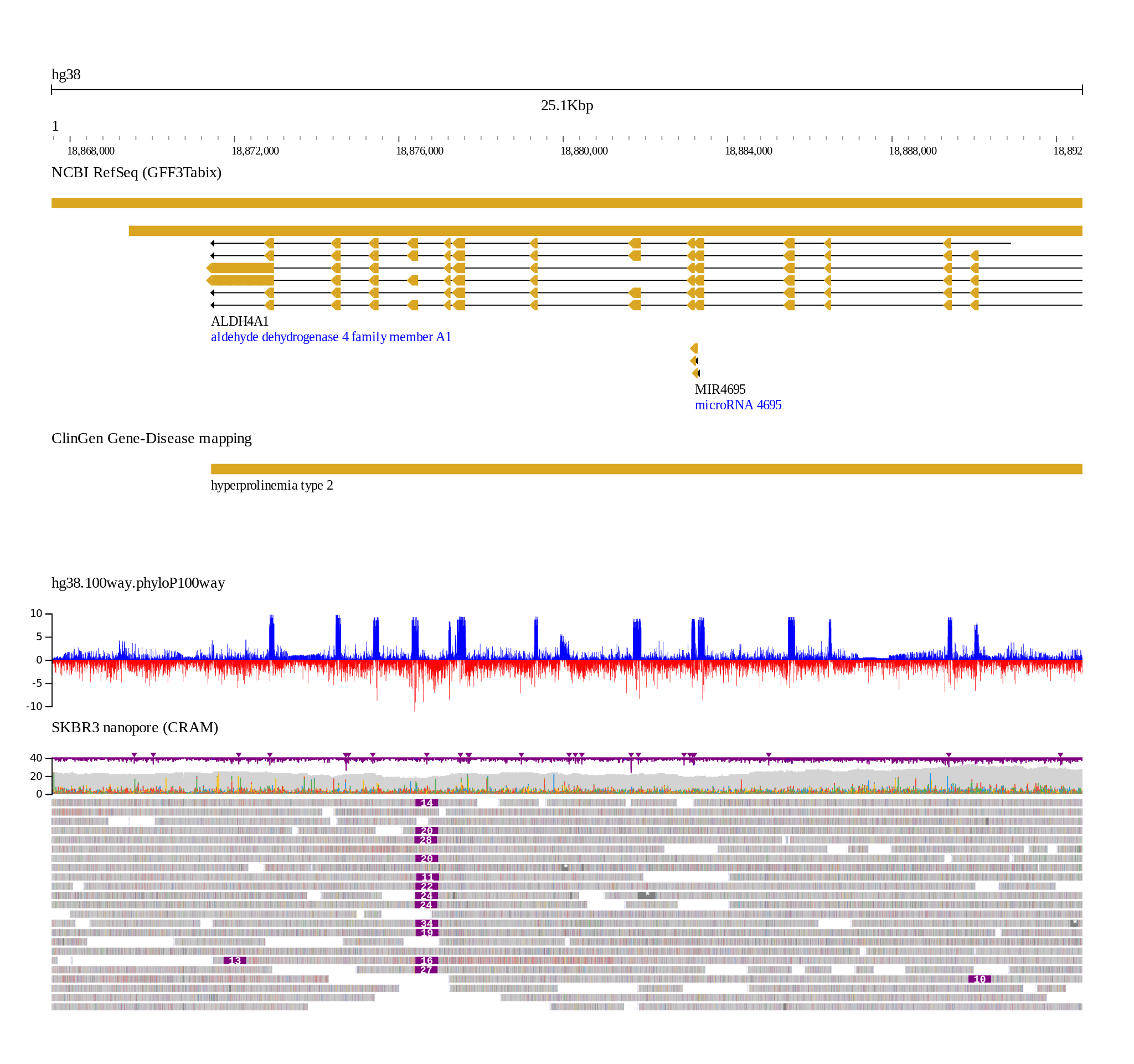
More examples EXAMPLES.md
Setup
You can install the @jbrowse/img package from npm, which, if your node is
configured in a typical configuration, will then have a command jb2export in
your path
npm install -g @jbrowse/img
If you are a developer and want to modify the code, see
developer guide for details
Example usages
Use with local files
We can call this script on local files, and it doesn't require a web browser,
not even a headless webbrowser, it just runs a node script and React SSR is used
to create the SVG
samtools faidx yourfile.fa
samtools index yourfile.bam
jb2export --fasta yourfile.fa --bam yourfile.bam --loc chr1:1,000,000-1,001,000 --out file.svg
If --out is not specified it writes to out.svg
Generate PNG instead of SVG
Supply a file with the png extension to --out, uses rsvg-convert so you will
need to install rsvg-convert to your system e.g. with
sudo apt install librsvg2-bin
jb2export --fasta yourfile.fa --bam yourfile.bam --loc chr1:1,000,000-1,001,000 --out file.png
Generate PDF instead of SVG
Supply a file with the pdf extension to --out, uses rsvg-convert so you will
need to install rsvg-convert to your system e.g. with
sudo apt install librsvg2-bin
jb2export --fasta yourfile.fa --bam yourfile.bam --loc chr1:1,000,000-1,001,000 --out file.pdf
Use with remote files
This example shows using remote files, e.g. with human hg19 and several tracks
Note the use of --aliases, which smoothes over refname differences e.g. fasta
contains 1 for chr1, and bigbed contains chr1, gff contains NC_000001.10
jb2export --fasta https://jbrowse.org/genomes/hg19/fasta/hg19.fa.gz \
--aliases https://jbrowse.org/genomes/hg19/hg19_aliases.txt \
--bigbed https://hgdownload.soe.ucsc.edu/gbdb/hg19/bbi/clinvar/clinvarMain.bb \
--gffgz https://jbrowse.org/genomes/hg19/ncbi_refseq/GRCh37_latest_genomic.sort.gff.gz \
--bigwig https://jbrowse.org/genomes/hg19/reads_lr_skbr3.fa_ngmlr-0.2.3_mapped.bam.regions.bw \
--loc 1:48,683,542..48,907,531
Customizing track
In addition to possibly specifying custom track configuration files, sometimes
specializing specifically track state is helpful. This example helps color and
sort by the read group (RG) tag
jb2export --fasta data/volvox/volvox.fa \
--bam data/volvox/volvox-rg.bam color:tag:RG sort:tag:RG height:400 \
--loc ctgA:609..968
You can see that instead of adding extra dash dash --flags, it is a colon based
syntax that follows a track definition.
The color and sort are specific to pileup, and height can apply to any track.
More options may be described here soon
Force render a large region
Some jbrowse track types (alignments, gene tracks, etc) will not display if
zoomed too far out. Add force:true to make it render
jb2export --bam file.bam force:true --loc 1:1,100,000-1,200,000 --fasta hg19.fa
Render only the SNPCoverage track of an alignments track
Renders only the snpcov subtrack at height 600 for file.bam
jb2export --bam file.bam snpcov height:600
Render the sequence track
If you are using the fasta argument, the refseq will be named "refseq" and can
be specified with the --configtracks tag. If you are using a pre-loaded
config.json then you can find the trackId to pass to --configtracks in there
jb2export --fasta data/volvox/volvox.fa --configtracks refseq --loc ctgA:1-100
Use with a jbrowse config.json (remote files in the config.json)
A config.json can be specified, and then we just refer to trackIds in this file,
and extra tracks can also be supplied that are outside of the config e.g. with
--bam
jb2export --config data/config.json \
--assembly hg19 \
--configtracks hg00096_highcov clinvar_cnv_hg19 \
--bam custom_bam.bam \
--loc 1:1,000,000-1,100,000
Respects the order of the files you input
Example:
jb2export --bam file1.bam --bigwig file.bw --bam file2.bam
This will respect the order of the tracks and list file1.bam, file.bw, and
file2.bam in that order. This requires us to use a custom command line parser
instead of an off-the-shelf one like yargs
Use a session file exported from jbrowse
If you use jbrowse-web, you can select File->Export session which produces a
session.json file, and then use the --session parameter. Make sure to specify
the assembly also, it currently does not infer the assembly from the session
jb2export --config data/skbr3/config.json \
--session session.json \
--assembly hg19
Plot whole-genome overview of bigwig
The special flag --loc all shows the full assembly, and there are a number of
custom bigwig plotting options that can help draw the bigwig genome wide
Example with logscale, manual setting of minmax score
jb2export --loc all \
--bigwig coverage.bw scaletype:log fill:false resolution:superfine height:400 color:purple minmax:1:1024 \
--assembly hg19 \
--config data/config.json
Example with linearscale, autoscore adjusted to "localsd" or mean plus/minus
three standard deviations
jb2export --loc all \
--bigwig coverage.bw autoscale:localsd fill:false resolution:superfine height:400 color:purple \
--assembly hg19 \
--config data/config.json
Use with a jbrowse config.json (local files in the config.json)
The jbrowse CLI tool (e.g. npm install -g @jbrowse/cli) refers to "uri" paths by
default, but you replace them with localPath like this
"vcfGzLocation": {
"uri": "volvox.dup.vcf.gz"
},
"vcfGzLocation": {
"localPath": "volvox.dup.vcf.gz"
}
Then you can call it like above
jb2export --config data/volvox/config.json \
--assembly volvox \
--configtracks volvox_sv \
--loc ctgA:1-50,000
The localPaths will be resolved relative to the file that is supplied so in this
example we would resolve data/volvox/volvox.dup.vcf.gz if "localPath":
"volvox.dup.vcf.gz" is used, and --config data/volvox/config.json is passed
See data/volvox/config.json for a config that contains localPaths, or
data/config.json for a config that just contains URLs
Parameters
Assembly params
- --fasta - filename or http(s) URL for a indexed or bgzip indexed FASTA file
- --aliases - tab separated "refName aliases" with column 1 matching the FASTA,
and other columns being aliases
Track params
Specify these with a filename (local to the computer) or a http(s) URL. Can
specify it multiple times e.g. --bam file1.bam --bam file2.bam
- --bigbed
- --gffgz
- --bedgz
- --vcfgz
- --bigwig
- --bam
- --cram
- --hic (wip)
Config file params (optional)
- --assembly - path to a JSON file containing a jbrowse 2 assembly config e.g.
data/assembly.json, can be used in place of --fasta
- --tracks - path to a JSON file containing a list of jbrowse 2 track configs
e.g. data/tracks.json
- --session - path to a JSON file containing a jbrowse 2 session config e.g.
data/session.json
- --config - path to a JSON file containing a full jbrowse 2 config e.g.
data/config.json
Other
- --loc - a locstring to navigate to
- --out - file to write the svg to
- --noRasterize - the canvas based tracks such as wiggle, read pileups, and hic
are rasterized to a PNG inside the svg by default. if you want it in all SVG
then use this flag but note that filesize may be much larger
Use --help
jb2export --help
jb2export [command]
Commands:
jb2export jb2export Creates a jbrowse 2 image snapshot
Options:
--version Show version number [boolean]
--config Path to config file [string]
--session Path to session file [string]
--assembly Path to an assembly configuration, or a name of an
assembly in the configFile [string]
--tracks Path to tracks portion of a session [string]
--loc A locstring to navigate to, or --loc all to view the
whole genome [string]
--fasta Supply a fasta for the assembly [string]
--aliases Supply aliases for the assembly, e.g. mapping of 1 to
chr1. Tab separated file where column 1 matches the
names from the FASTA [string]
-w, --width Set the width of the svg canvas, default 1500px [number]
--configtracks A list of track labels from a config file [array]
--bam A bam file, flag --bam can be used multiple times to
specify multiple bam files [array]
--bigwig A bigwig file, the --bigwig flag can be used multiple
times to specify multiple bigwig files [array]
--cram A cram file, the --cram flag can be used multiple times
to specify multiple cram files [array]
--vcfgz A tabixed VCF, the --vcfgz flag can be used multiple
times to specify multiple vcfgz files [array]
--gffgz A tabixed GFF, the --gffgz can be used multiple times to
specify multiple gffgz files [array]
--hic A .hic file, the --hic can be used multiple times to
specify multiple hic files [array]
--bigbed A .bigBed file, the --bigbed can be used multiple times
to specify multiple bigbed files [array]
--bedgz A bed tabix file, the --bedgz can be used multiple times
to specify multiple bedtabix files [array]
--out File to output to. Default: out.svg
[string] [default: "out.svg"]
--noRasterize Use full SVG rendering with no rasterized layers, this
can substantially increase filesize [boolean]
--defaultSession Use the defaultSession from config.json [boolean]
-h, --help Show help [boolean]
Convert to PNG
The tool will automatically try to run rsvg-convert and convert to PNG if a
filename with png extension is supplied to --out
Alternatively, you can do so manually with commands like this
sudo apt install inkscape
inkscape --export-type png --export-filename out.png -w 2048 out.svg
sudo apt install librsvg2-bin
rsvg-convert -w 2048 out.svg -o out.png
sudo apt install imagemagick
convert -size 2048x out.svg out.png
Troubleshooting
I don't get any outputted svg and no message
The error reporting from the app is not very good at the moment so often has
silent failures. Confirm that your fasta file to your pass to --fasta is indexed
in this case e.g. samtools faidx yourfile.fa so that your have a
yourfile.fa.fai alongside yourfile.fa
I get a lot of warnings during npm install -g @jbrowse/img
There are some new features in the latest NPM (2021, v7) related to
peerDependencies that may produce some warnings. It should work even despite
making warnings, but you can use yarn to install or use legacy peer dependencies
if you want to avoid install time warningsvg