ipyspeck
ipyspeck Stats
| Latest Release |

|
| PyPI Downloads |


|
Speck
Speck is a molecule renderer with the goal of producing figures that are as attractive as they are practical. Express your molecule clearly and with style.
ipypeck
Ipyspeck is a ipywidget wrapping speck to be used on a Jupyter notebook as a regular widget.
Usage
The ipyspeck widget renders xyz molecules.
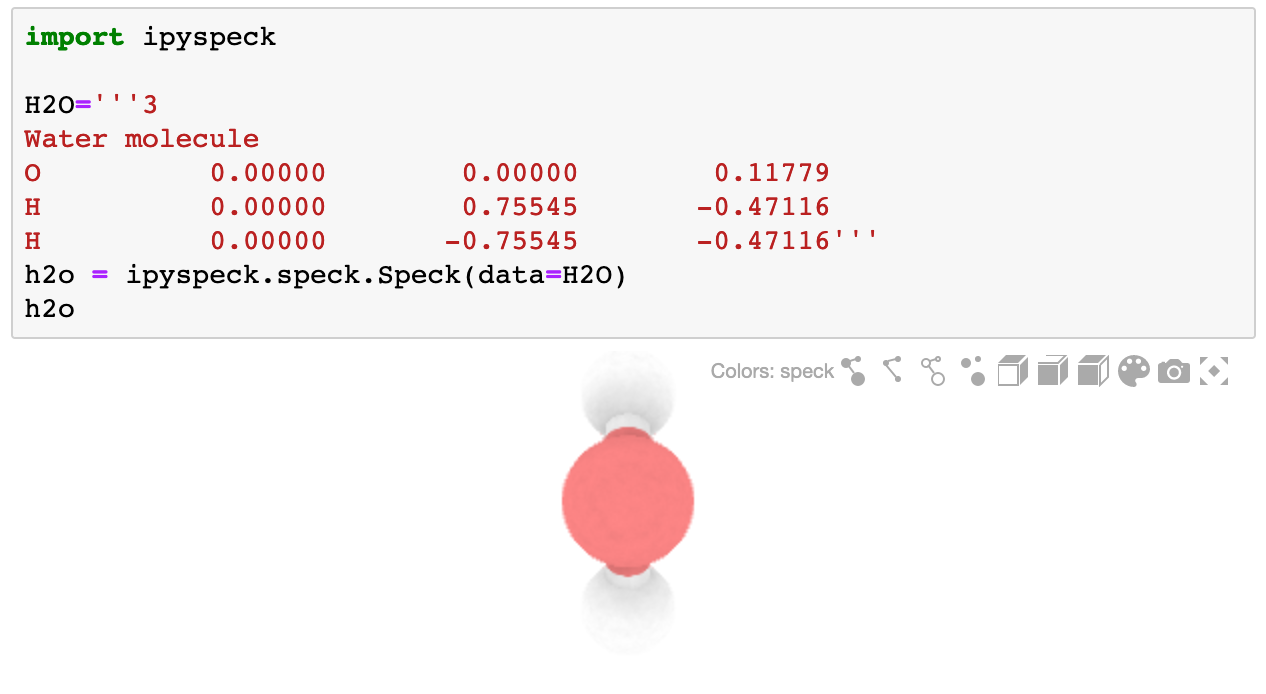
import ipyspeck
H2O='''3
Water molecule
O 0.00000 0.00000 0.11779
H 0.00000 0.75545 -0.47116
H 0.00000 -0.75545 -0.47116'''
h2o = ipyspeck.speck.Speck(data=H2O)
h2o
Ideally it should be used as part of a container widget (such as Box, VBox, Grid, ...)
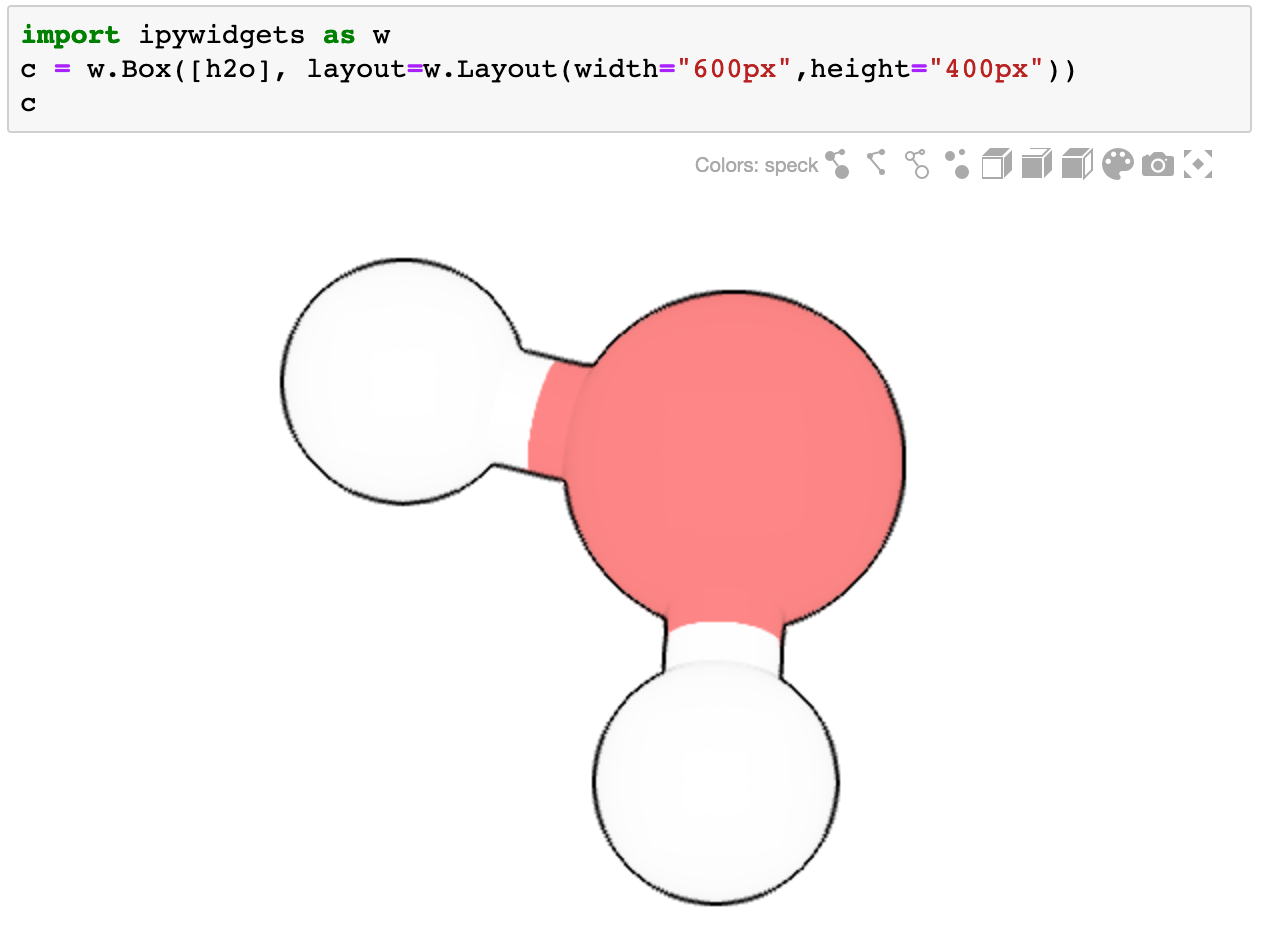
import ipywidgets as w
c = w.Box([h2o], layout=w.Layout(width="600px",height="400px"))
c
The visualization parameters can be modified
h2o.atomScale = 0.3
h2o.bondScale = 0.3
h2o.outline = 0
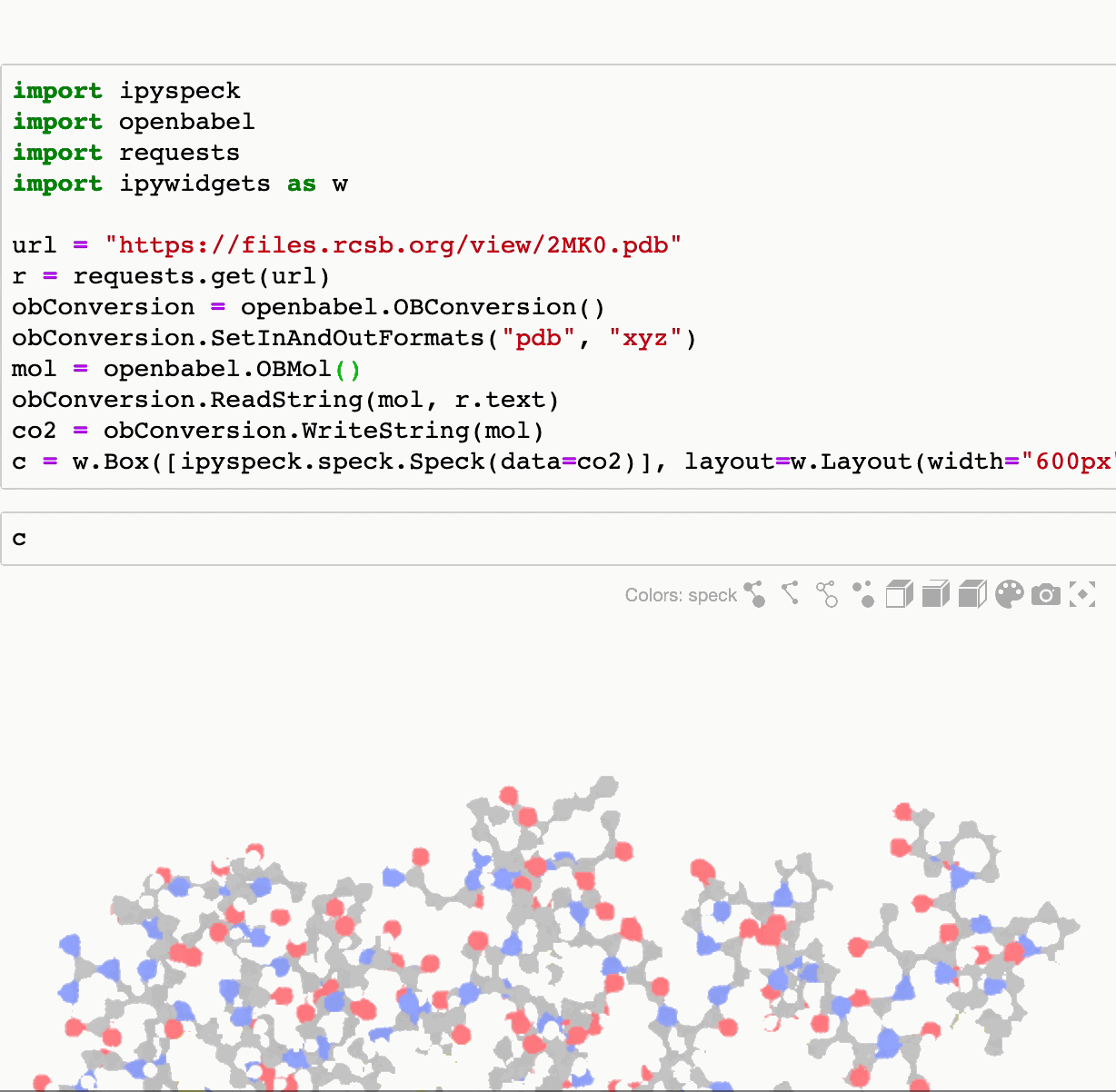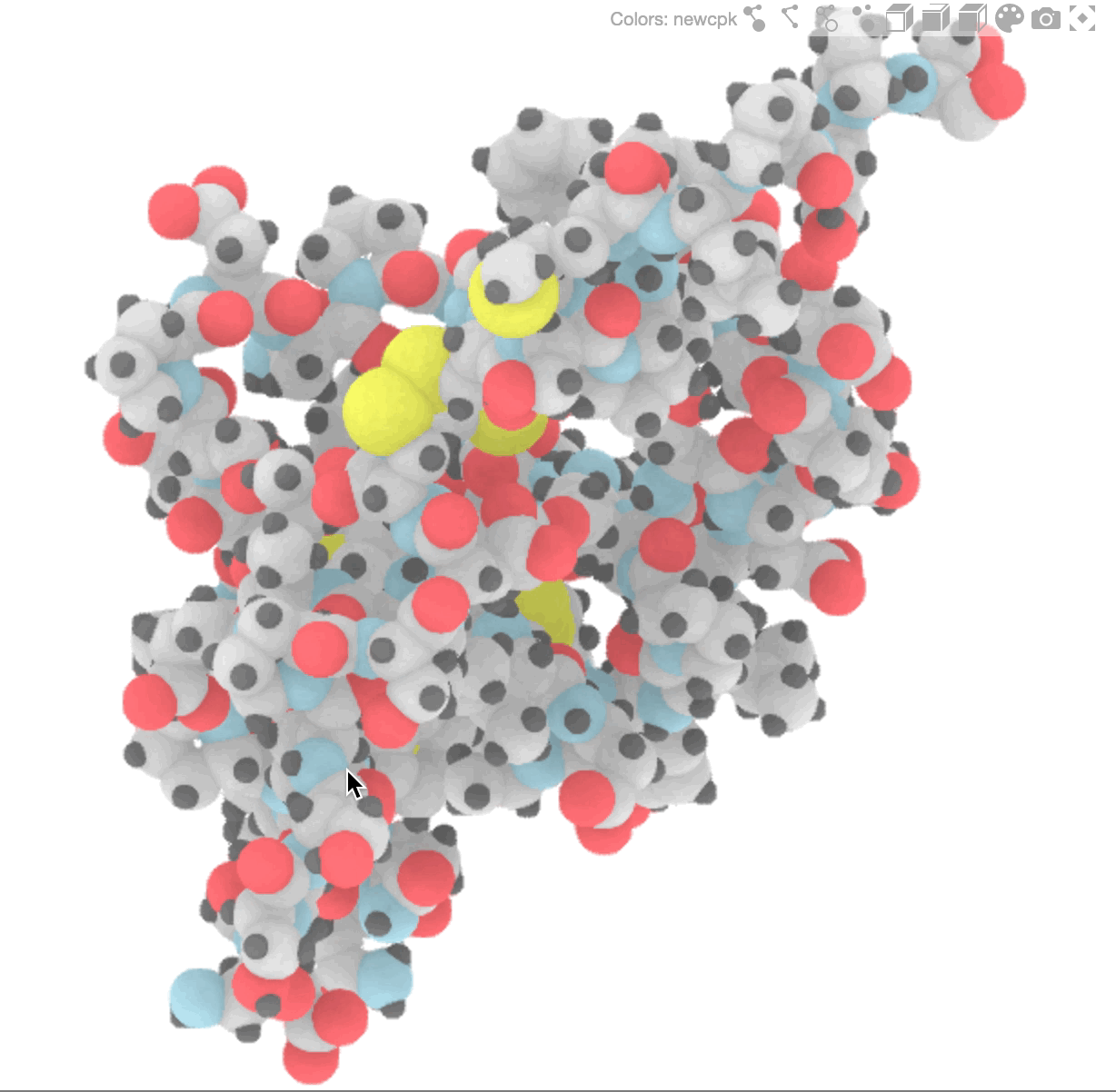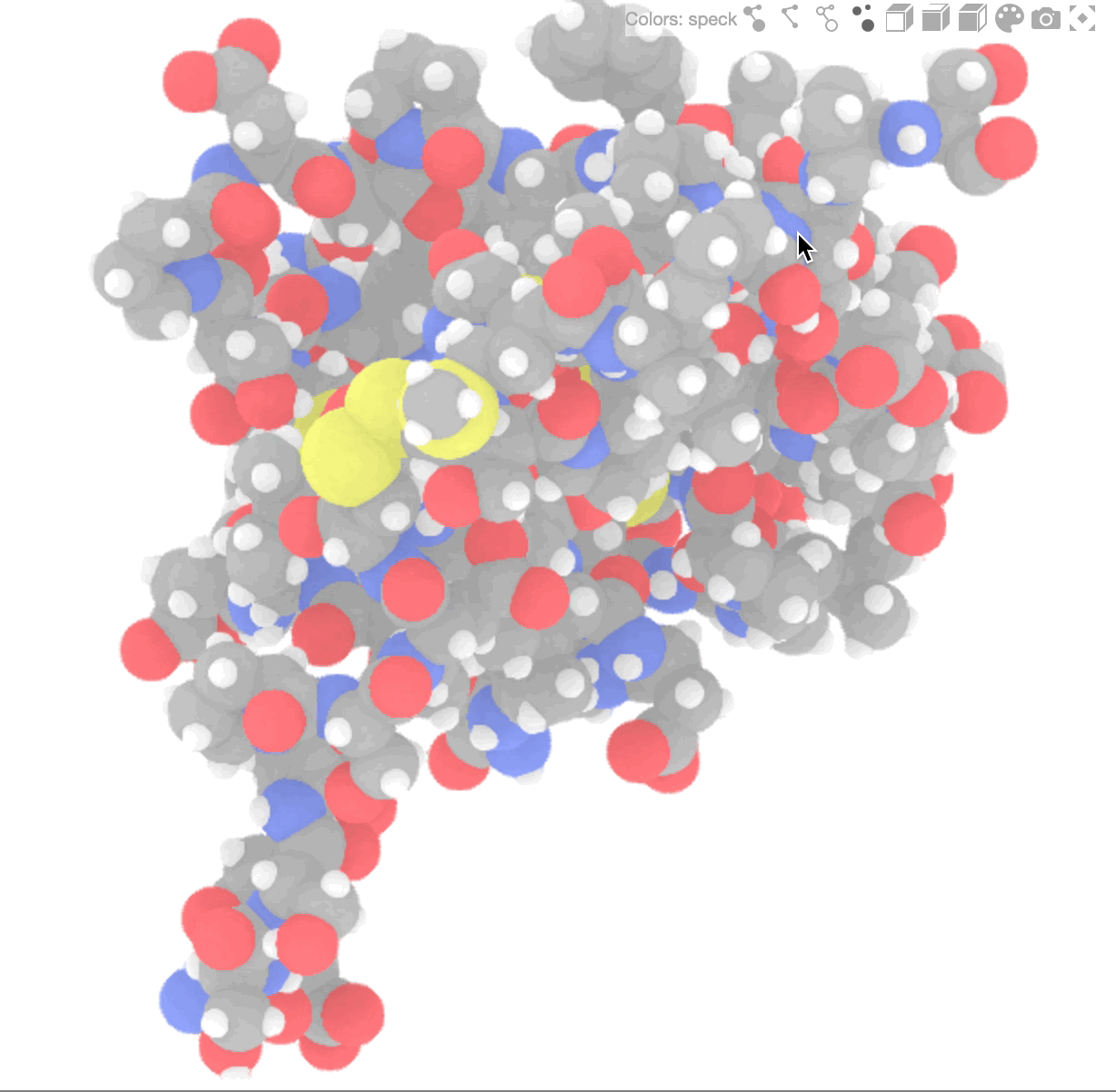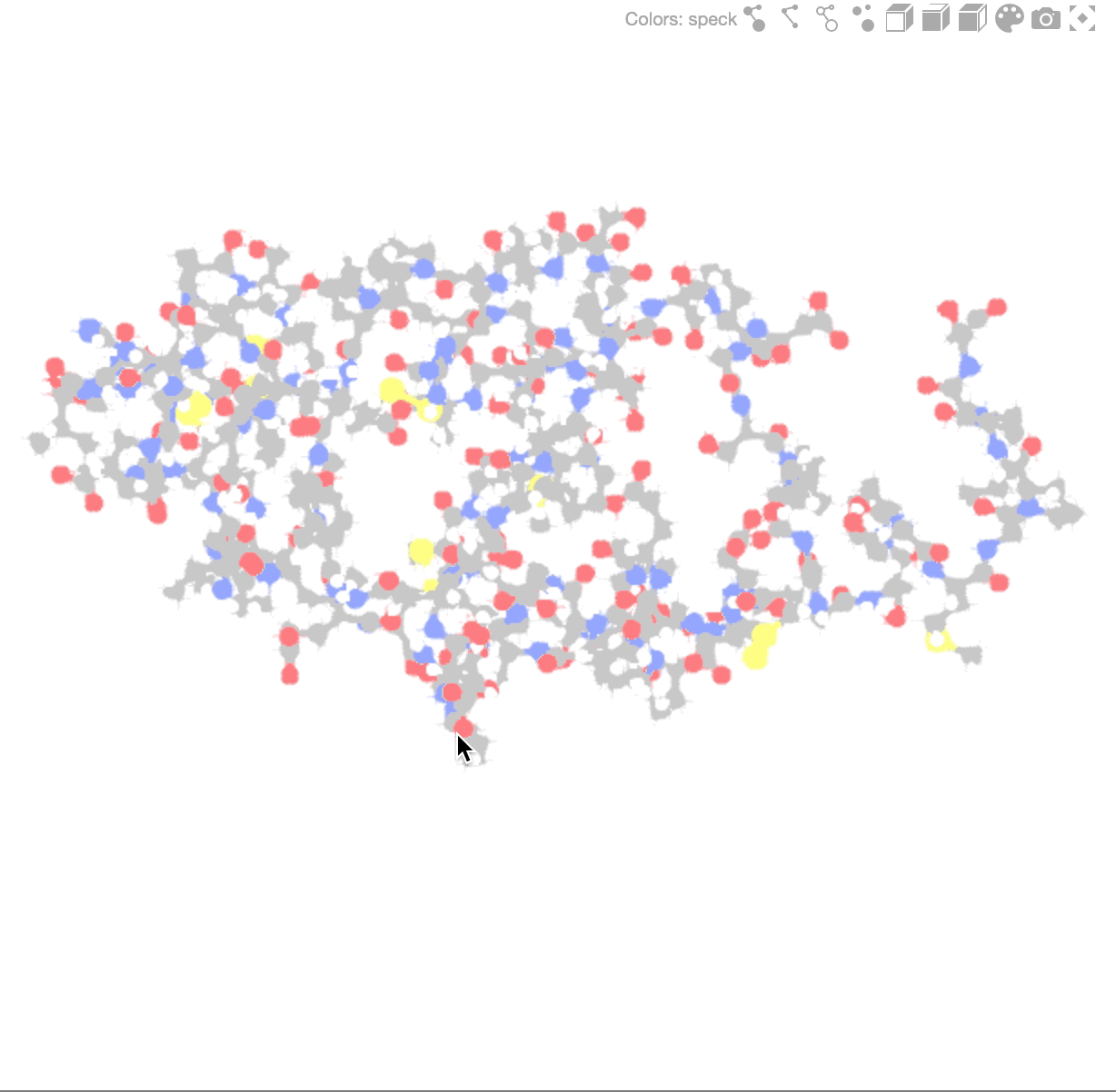
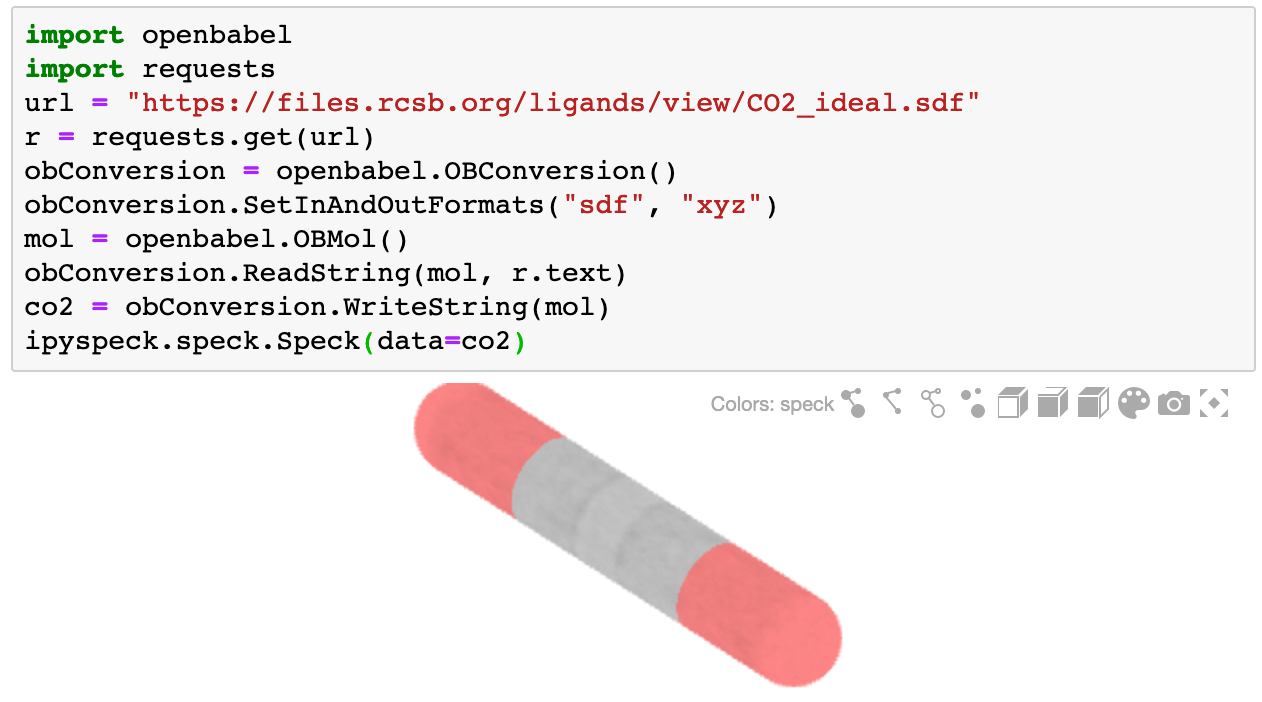
To render molecules on different formats openbabel can be used to translate them as xyz
import openbabel
import requests
url = "https://files.rcsb.org/ligands/view/CO2_ideal.sdf"
r = requests.get(url)
obConversion = openbabel.OBConversion()
obConversion.SetInAndOutFormats("sdf", "xyz")
mol = openbabel.OBMol()
obConversion.ReadString(mol, r.text)
co2 = obConversion.WriteString(mol)
ipyspeck.speck.Speck(data=co2)
Installation
To install use pip:
$ pip install ipyspeck
$ jupyter nbextension enable --py --sys-prefix ipyspeck
To install for jupyterlab
$ jupyter labextension install ipyspeck
For a development installation (requires npm),
$ git clone https://github.com//denphi//speck.git
$ cd speck/widget/ipyspeck
$ pip install -e .
$ jupyter nbextension install --py --symlink --sys-prefix ipyspeck
$ jupyter nbextension enable --py --sys-prefix ipyspeck
$ jupyter labextension install js
When actively developing your extension, build Jupyter Lab with the command:
$ jupyter lab --watch
This takes a minute or so to get started, but then automatically rebuilds JupyterLab when your javascript changes.
Note on first jupyter lab --watch, you may need to touch a file to get Jupyter Lab to open.
Gallery