
Research
Security News
Malicious npm Packages Inject SSH Backdoors via Typosquatted Libraries
Socket’s threat research team has detected six malicious npm packages typosquatting popular libraries to insert SSH backdoors.
💻 See the Installation page in the documentation for detailed instructions.
📚 A lot of documentation is available at https://AdaptiveMotorControlLab.github.io/CellSeg3D
You can also generate docs by running make html in the docs/ folder.
To use the plugin, please run:
napari
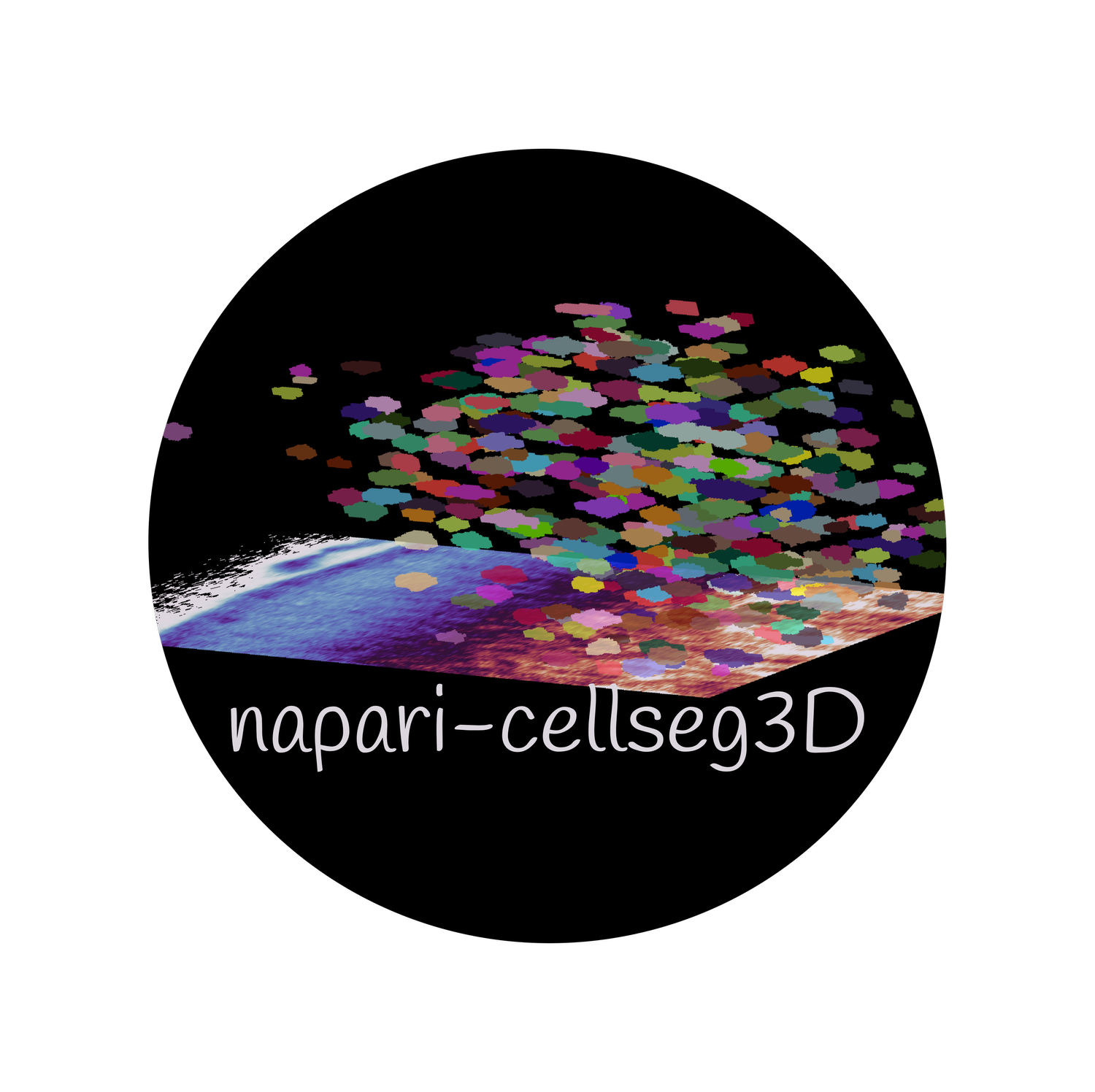
Then go into Plugins > napari-cellseg3d, and choose which tool to use.
New version : v0.2.0
Previous additions :
To avoid issues when installing on the ARM64 architecture, please follow these steps.
Create a new conda env using the provided conda/napari_CellSeg3D_ARM64.yml file :
git clone https://github.com/AdaptiveMotorControlLab/CellSeg3d.git
cd CellSeg3d
conda env create -f conda/CellSeg3D_ARM64.yml
conda activate napari_CellSeg3D_ARM64
Install a Qt backend (PySide or PyQt5)
Launch napari, the plugin should be available in the plugins menu.
Python 3.8 or 3.9 required. Requires napari, PyTorch and MONAI. Compatible with Windows, MacOS and Linux. Installation should not take more than 30 minutes, depending on your internet connection.
For PyTorch, please see the PyTorch website for installation instructions.
A CUDA-capable GPU is not needed but very strongly recommended, especially for training.
If you get errors from MONAI regarding missing readers, please see MONAI's optional dependencies page for instructions on getting the readers required by your images.
After installation, you can run the plugin by running:
napari
and launching the plugin from the Plugins menu.
You may use the test volume in the examples folder to test the inference and review tools.
This should run in far less than five minutes on a modern computer.
You may also find a demo Colab notebook in the notebooks folder.
Help us make the code better by reporting issues and adding your feature requests!
If you encounter any problems, please file an issue along with a detailed description.
Before testing, install all requirements using pip install napari-cellseg3d[test].
pydensecrf is also required for testing.
To run tests locally:
pytest in the plugin foldercoverage run --source=napari_cellseg3d -m pytest then coverage xml to generate a .xml coverage file.tox in the plugin folder (will simulate tests with several python and OS configs, requires substantial storage space)Contributions are very welcome.
Please ensure the coverage at least stays the same before you submit a pull request.
For local installation from Github cloning, please run:
pip install -e .
Distributed under the terms of the MIT license.
"napari-cellseg3d" is free and open source software.
This plugin was developed by originally Cyril Achard, Maxime Vidal, Mackenzie Mathis. This work was funded, in part, from the Wyss Center to the Mathis Laboratory of Adaptive Intelligence. Please refer to the documentation for full acknowledgements.
This napari plugin was generated with Cookiecutter using @napari's cookiecutter-napari-plugin template.
FAQs
Plugin for cell segmentation in 3D
We found that napari-cellseg3d demonstrated a healthy version release cadence and project activity because the last version was released less than a year ago. It has 2 open source maintainers collaborating on the project.
Did you know?

Socket for GitHub automatically highlights issues in each pull request and monitors the health of all your open source dependencies. Discover the contents of your packages and block harmful activity before you install or update your dependencies.

Research
Security News
Socket’s threat research team has detected six malicious npm packages typosquatting popular libraries to insert SSH backdoors.

Security News
MITRE's 2024 CWE Top 25 highlights critical software vulnerabilities like XSS, SQL Injection, and CSRF, reflecting shifts due to a refined ranking methodology.

Security News
In this segment of the Risky Business podcast, Feross Aboukhadijeh and Patrick Gray discuss the challenges of tracking malware discovered in open source softare.