
Security News
/Research
Wallet-Draining npm Package Impersonates Nodemailer to Hijack Crypto Transactions
Malicious npm package impersonates Nodemailer and drains wallets by hijacking crypto transactions across multiple blockchains.
The bioinfokit toolkit aimed to provide various easy-to-use functionalities to analyze,
visualize, and interpret the biological data generated from genome-scale omics experiments.
How to install:
bioinfokit requires
bioinfokit can be installed using pip, easy_install and git.
Install using pip for Python 3 (easiest way)
# install
pip install bioinfokit
# upgrade to latest version
pip install bioinfokit --upgrade
# uninstall
pip uninstall bioinfokit
Install using easy_install for Python 3 (easiest way)
# install latest version
easy_install bioinfokit
# specific version
easy_install bioinfokit==0.3
# uninstall
pip uninstall bioinfokit
Install using conda
conda install -c bioconda bioinfokit
Install using git
# download and install bioinfokit (Tested on Linux, Mac, Windows)
git clone https://github.com/reneshbedre/bioinfokit.git
cd bioinfokit
python setup.py install
>>> import bioinfokit
>>> bioinfokit.__version__
'0.4'
If you enjoy bioinfokit, consider supporting me,
latest update v2.0.8
bioinfokit.visuz.GeneExpression.volcano(df, lfc, pv, lfc_thr, pv_thr, color, valpha, geneid, genenames, gfont, dim, r, ar, dotsize, markerdot, sign_line, gstyle, show, figtype, axtickfontsize, axtickfontname, axlabelfontsize, axlabelfontname, axxlabel, axylabel, xlm, ylm, plotlegend, legendpos, figname, legendanchor, legendlabels, theme)
| Parameters | Description |
|---|---|
df | Pandas dataframe table having atleast gene IDs, log fold change, P-values or adjusted P-values columns |
lfc | Name of a column having log or absolute fold change values [string][default:logFC] |
pv | Name of a column having P-values or adjusted P-values [string][default:p_values] |
lfc_thr | Log fold change cutoff for up and downregulated genes [Tuple or list][default:(1.0, 1.0)] |
pv_thr | p value or adjusted p value cutoff for up and downregulated genes [Tuple or list][default:(0.05, 0.05)] |
color | Tuple of three colors [Tuple or list][default: color=("green", "grey", "red")] |
valpha | Transparency of points on volcano plot [float (between 0 and 1)][default: 1.0] |
geneid | Name of a column having gene Ids. This is necessary for plotting gene label on the points [string][default: None] |
genenames | Tuple of gene Ids to label the points. The gene Ids must be present in the geneid column. If this option set to "deg" it will label all genes defined by lfc_thr and pv_thr [string, tuple, dict][default: None] |
gfont | Font size for genenames [float][default: 10.0]. gfont not compatible with gstyle=2. |
dim | Figure size [Tuple of two floats (width, height) in inches][default: (5, 5)] |
r | Figure resolution in dpi [int][default: 300]. Not compatible with show= True |
ar | Rotation of X and Y-axis ticks labels [float][default: 90] |
dotsize | The size of the dots in the plot [float][default: 8] |
markerdot | Shape of the dot marker. See more options at https://matplotlib.org/3.1.1/api/markers_api.html [string][default: "o"] |
sign_line | Show grid lines on plot with defined log fold change (lfc_thr) and P-value (pv_thr) threshold value [True or False][default:False] |
gstyle | Style of the text for genenames. 1 for default text and 2 for box text [int][default: 1] |
show | Show the figure on console instead of saving in current folder [True or False][default:False] |
figtype | Format of figure to save. Supported format are eps, pdf, pgf, png, ps, raw, rgba, svg, svgz [string][default:'png'] |
axtickfontsize | Font size for axis ticks [float][default: 9] |
axtickfontname | Font name for axis ticks [string][default: 'Arial'] |
axlabelfontsize | Font size for axis labels [float][default: 9] |
axlabelfontname | Font name for axis labels [string][default: 'Arial'] |
axxlabel | Label for X-axis. If you provide this option, default label will be replaced [string][default: None] |
axylabel | Label for Y-axis. If you provide this option, default label will be replaced [string][default: None] |
xlm | Range of ticks to plot on X-axis [float (left, right, interval)][default: None] |
ylm | Range of ticks to plot on Y-axis [float (bottom, top, interval)][default: None] |
plotlegend | plot legend on volcano plot [True or False][default:False] |
legendpos | position of the legend on plot. For more options see loc parameter at https://matplotlib.org/3.1.1/api/_as_gen/matplotlib.pyplot.legend.html [string ][default:"best"] |
figname | name of figure [string ][default:"volcano"] |
legendanchor | position of the legend outside of the plot. For more options see bbox_to_anchor parameter at https://matplotlib.org/3.1.1/api/_as_gen/matplotlib.pyplot.legend.html [list][default:None] |
legendlabels | legend label names. If you provide custom label names keep the same order of label names as default [list][default:['significant up', 'not significant', 'significant down']] |
theme | Change background theme. If theme set to dark, the dark background will be produced instead of white [string][default:'None'] |
Returns:
Volcano plot image in same directory (volcano.png) Working example
latest update v2.0.8
bioinfokit.visuz.GeneExpression.involcano(table, lfc, pv, lfc_thr, pv_thr, color, valpha, geneid, genenames, gfont, gstyle, dotsize, markerdot, r, dim, show, figtype, axxlabel, axylabel, axlabelfontsize, axtickfontsize, axtickfontname, plotlegend, legendpos, legendanchor, figname, legendlabels, ar, theme)
| Parameters | Description |
|---|---|
table | Pandas dataframe table having atleast gene IDs, log fold change, P-values or adjusted P-values |
lfc | Name of a column having log fold change values [default:logFC] |
pv | Name of a column having P-values or adjusted P-values [default:p_values] |
lfc_thr | Log fold change cutoff for up and downregulated genes [Tuple or list] [default:(1.0, 1.0)] |
pv_thr | p value or adjusted p value cutoff for up and downregulated genes [Tuple or list] [default:(0.05, 0.05)] |
color | Tuple of three colors [Tuple or list][default: color=("green", "grey", "red")] |
valpha | Transparency of points on volcano plot [float (between 0 and 1)][default: 1.0] |
geneid | Name of a column having gene Ids. This is necessary for plotting gene label on the points [string][default: None] |
genenames | Tuple of gene Ids to label the points. The gene Ids must be present in the geneid column. If this option set to "deg" it will label all genes defined by lfc_thr and pv_thr [string, Tuple, dict][default: None] |
gfont | Font size for genenames [float][default: 10.0] |
gstyle | Style of the text for genenames. 1 for default text and 2 for box text [int][default: 1] |
dotsize | The size of the dots in the plot [float][default: 8] |
markerdot | Shape of the dot marker. See more options at https://matplotlib.org/3.1.1/api/markers_api.html [string][default: "o"] |
dim | Figure size [Tuple of two floats (width, height) in inches][default: (5, 5)] |
r | Figure resolution in dpi [int][default: 300]. Not compatible with show= True |
figtype | Format of figure to save. Supported format are eps, pdf, pgf, png, ps, raw, rgba, svg, svgz [string][default:'png'] |
show | Show the figure on console instead of saving in current folder [True or False][default:False] |
axxlabel | Label for X-axis. If you provide this option, default label will be replaced [string][default: None] |
axylabel | Label for Y-axis. If you provide this option, default label will be replaced [string][default: None] |
axlabelfontsize | Font size for axis labels [float][default: 9] |
axtickfontsize | Font size for axis ticks [float][default: 9] |
axtickfontname | Font name for axis ticks [string][default: 'Arial'] |
plotlegend | plot legend on inverted volcano plot [True or False][default:False] |
legendpos | position of the legend on plot. For more options see loc parameter at https://matplotlib.org/3.1.1/api/_as_gen/matplotlib.pyplot.legend.html [string ][default:"best"] |
legendanchor | position of the legend outside of the plot. For more options see bbox_to_anchor parameter at https://matplotlib.org/3.1.1/api/_as_gen/matplotlib.pyplot.legend.html [list][default:None] |
figname | name of figure [string ][default:"involcano"] |
legendlabels | legend label names. If you provide custom label names keep the same order of label names as default [list][default:['significant up', 'not significant', 'significant down']] |
ar | Rotation of X and Y-axis ticks labels [float][default: 90] |
theme | Change background theme. If theme set to dark, the dark background will be produced instead of white [string][default:'None'] |
Returns:
Inverted volcano plot image in same directory (involcano.png) Working example
latest update v2.0.7
bioinfokit.visuz.GeneExpression.ma(df, lfc, ct_count, st_count, pv, basemean, lfc_thr, color, dim, dotsize, show, r, valpha, figtype, axxlabel, axylabel, axlabelfontsize, axtickfontsize, axtickfontname, xlm, ylm, fclines, fclinescolor, legendpos, legendanchor, figname, legendlabels, plotlegend, ar, theme, geneid, genenames, gfont, gstyle, title)
| Parameters | Description |
|---|---|
df | Pandas dataframe table having atleast gene IDs, log fold change, and normalized counts (control and treatment) columns |
lfc | Name of a column having log fold change values [default:"logFC"] |
ct_count | Name of a column having count values for control sample.Ignored if basemean provided [default:"value1"] |
st_count | Name of a column having count values for treatment sample. Ignored if basemean provided [default:"value2"] |
pv | Name of a column having p values or adjusted p values |
basemean | Basemean (mean of normalized counts) from DESeq2 results |
lfc_thr | Log fold change cutoff for up and downregulated genes [Tuple or list][default:(1.0, 1.0)] |
color | Tuple of three colors [Tuple or list][default: ("green", "grey", "red")] |
dotsize | The size of the dots in the plot [float][default: 8] |
markerdot | Shape of the dot marker. See more options at https://matplotlib.org/3.1.1/api/markers_api.html [string][default: "o"] |
valpha | Transparency of points on plot [float (between 0 and 1)][default: 1.0] |
dim | Figure size [Tuple of two floats (width, height) in inches][default: (5, 5)] |
r | Figure resolution in dpi [int][default: 300]. Not compatible with show= True |
figtype | Format of figure to save. Supported format are eps, pdf, pgf, png, ps, raw, rgba, svg, svgz [string][default:'png'] |
show | Show the figure on console instead of saving in current folder [True or False][default:False] |
axxlabel | Label for X-axis. If you provide this option, default label will be replaced [string][default: None] |
axylabel | Label for Y-axis. If you provide this option, default label will be replaced [string][default: None] |
axlabelfontsize | Font size for axis labels [float][default: 9] |
axtickfontsize | Font size for axis ticks [float][default: 9] |
axtickfontname | Font name for axis ticks [string][default: 'Arial'] |
xlm | Range of ticks to plot on X-axis [float (left, right, interval)][default: None] |
ylm | Range of ticks to plot on Y-axis [float (bottom, top, interval)][default: None] |
fclines | draw log fold change threshold lines as defines by lfc [True or False][default:False] |
fclinescolor | color of fclines [string][default: '#2660a4'] |
plotlegend | plot legend on MA plot [True or False][default:False] |
legendpos | position of the legend on plot. For more options see loc parameter at https://matplotlib.org/3.1.1/api/_as_gen/matplotlib.pyplot.legend.html [string ][default:"best"] |
legendanchor | position of the legend outside of the plot. For more options see bbox_to_anchor parameter at https://matplotlib.org/3.1.1/api/_as_gen/matplotlib.pyplot.legend.html [list][default:None] |
figname | name of figure [string ][default:"ma"] |
legendlabels | legend label names. If you provide custom label names keep the same order of label names as default [list][default:['significant up', 'not significant', 'significant down']] |
ar | Rotation of X and Y-axis ticks labels [float][default: 90] |
theme | Change background theme. If theme set to dark_background, the dark background will be produced instead of default white. See more themes here [string][default:'None'] |
geneid | Name of a column having gene Ids. This is necessary for plotting gene label on the points [string][default: None] |
genenames | Tuple of gene Ids to label the points. The gene Ids must be present in the geneid column. If this option set to "deg" it will label all genes defined by lfc_thr and pv_thr [string, Tuple, dict][default: None] |
gfont | Font size for genenames [float][default: 10.0] |
gstyle | Style of the text for genenames. 1 for default text and 2 for box text [int][default: 1] |
title | Add main title to the plot [string][default: None] |
Returns:
MA plot image in same directory (ma.png)
latest update v2.0.1
bioinfokit.visuz.gene_exp.hmap(table, cmap='seismic', scale=True, dim=(6, 8), rowclus=True, colclus=True, zscore=None, xlabel=True, ylabel=True, tickfont=(12, 12), show, r, figtype, figname, theme)
| Parameters | Description |
|---|---|
file | CSV delimited data file. It should not have NA or missing values |
cmap | Color Palette for heatmap [string][default: 'seismic'] |
scale | Draw a color key with heatmap [boolean (True or False)][default: True] |
dim | heatmap figure size [Tuple of two floats (width, height) in inches][default: (6, 8)] |
rowclus | Draw hierarchical clustering for rows [boolean (True or False)][default: True] |
colclus | Draw hierarchical clustering for columns [boolean (True or False)][default: True] |
zscore | Z-score standardization of row (0) or column (1). It works when clus is True. [None, 0, 1][default: None] |
xlabel | Plot X-label [boolean (True or False)][default: True] |
ylabel | Plot Y-label [boolean (True or False)][default: True] |
tickfont | Fontsize for X and Y-axis tick labels [Tuple of two floats][default: (14, 14)] |
show | Show the figure on console instead of saving in current folder [True or False][default:False] |
r | Figure resolution in dpi [int][default: 300]. Not compatible with show= True |
figtype | Format of figure to save. Supported format are eps, pdf, pgf, png, ps, raw, rgba, svg, svgz [string][default:'png'] |
figname | name of figure [string ][default:"heatmap"] |
theme | Change background theme. If theme set to dark, the dark background will be produced instead of white [string][default:'None'] |
Returns:
heatmap plot (heatmap.png, heatmap_clus.png)
latest update v2.0.1
bioinfokit.visuz.cluster.screeplot(obj, axlabelfontsize, axlabelfontname, axxlabel, axylabel, figtype, r, show, dim, theme)
| Parameters | Description |
|---|---|
obj | list of component name and component variance |
axlabelfontsize | Font size for axis labels [float][default: 9] |
axlabelfontname | Font name for axis labels [string][default: 'Arial'] |
axxlabel | Label for X-axis. If you provide this option, default label will be replaced [string][default: None] |
axylabel | Label for Y-axis. If you provide this option, default label will be replaced [string][default: None] |
figtype | Format of figure to save. Supported format are eps, pdf, pgf, png, ps, raw, rgba, svg, svgz [string][default:'png'] |
r | Figure resolution in dpi [int][default: 300] |
show | Show the figure on console instead of saving in current folder [True or False][default:False] |
dim | Figure size [Tuple of two floats (width, height) in inches][default: (6, 4)] |
theme | Change background theme. If theme set to dark, the dark background will be produced instead of white [string][default:'None'] |
Returns:
Scree plot image (screeplot.png will be saved in same directory)
latest update v2.0.1
bioinfokit.visuz.cluster.pcaplot(x, y, z, labels, var1, var2, var3, axlabelfontsize, axlabelfontname, figtype, r, show, plotlabels, dim, theme)
| Parameters | Description |
|---|---|
x | loadings (correlation coefficient) for principal component 1 (PC1) |
y | loadings (correlation coefficient) for principal component 2 (PC2) |
z | loadings (correlation coefficient) for principal component 3 (PC2) |
labels | original variables labels from dataframe used for PCA |
var1 | Proportion of PC1 variance [float (0 to 1)] |
var2 | Proportion of PC2 variance [float (0 to 1)] |
var3 | Proportion of PC3 variance [float (0 to 1)] |
axlabelfontsize | Font size for axis labels [float][default: 9] |
axlabelfontname | Font name for axis labels [string][default: 'Arial'] |
figtype | Format of figure to save. Supported format are eps, pdf, pgf, png, ps, raw, rgba, svg, svgz [string][default:'png'] |
r | Figure resolution in dpi [int][default: 300] |
show | Show the figure on console instead of saving in current folder [True or False][default:False] |
plotlabels | Plot labels as defined by labels parameter [True or False][default:True] |
dim | Figure size [Tuple of two floats (width, height) in inches][default: (6, 4)] |
theme | Change background theme. If theme set to dark, the dark background will be produced instead of white [string][default:'None'] |
Returns:
PCA loadings plot 2D and 3D image (pcaplot_2d.png and pcaplot_3d.png will be saved in same directory)
latest update v2.0.2
bioinfokit.visuz.cluster.biplot(cscore, loadings, labels, var1, var2, var3, axlabelfontsize, axlabelfontname, figtype, r, show, markerdot, dotsize, valphadot, colordot, arrowcolor, valphaarrow, arrowlinestyle, arrowlinewidth, centerlines, colorlist, legendpos, datapoints, dim, theme)
| Parameters | Description |
|---|---|
cscore | principal component scores (obtained from PCA().fit_transfrom() function in sklearn.decomposition) |
loadings | loadings (correlation coefficient) for principal components |
labels | original variables labels from dataframe used for PCA |
var1 | Proportion of PC1 variance [float (0 to 1)] |
var2 | Proportion of PC2 variance [float (0 to 1)] |
var3 | Proportion of PC3 variance [float (0 to 1)] |
axlabelfontsize | Font size for axis labels [float][default: 9] |
axlabelfontname | Font name for axis labels [string][default: 'Arial'] |
figtype | Format of figure to save. Supported format are eps, pdf, pgf, png, ps, raw, rgba, svg, svgz [string][default:'png'] |
r | Figure resolution in dpi [int][default: 300] |
show | Show the figure on console instead of saving in current folder [True or False][default:False] |
markerdot | Shape of the dot on plot. See more options at https://matplotlib.org/3.1.1/api/markers_api.html [string][default: "o"] |
dotsize | The size of the dots in the plot [float][default: 6] |
valphadot | Transparency of dots on plot [float (between 0 and 1)][default: 1] |
colordot | Color of dots on plot [string or list ][default:"#4a4e4d"] |
arrowcolor | Color of the arrow [string ][default:"#fe8a71"] |
valphaarrow | Transparency of the arrow [float (between 0 and 1)][default: 1] |
arrowlinestyle | line style of the arrow. check more styles at https://matplotlib.org/3.1.0/gallery/lines_bars_and_markers/linestyles.html [string][default: '-'] |
arrowlinewidth | line width of the arrow [float][default: 1.0] |
centerlines | draw center lines at x=0 and y=0 for 2D plot [bool (True or False)][default: True] |
colorlist | list of the categories to assign the color [list][default:None] |
legendpos | position of the legend on plot. For more options see loc parameter at https://matplotlib.org/3.1.1/api/_as_gen/matplotlib.pyplot.legend.html [string ][default:"best"] |
datapoints | plot data points on graph [bool (True or False)][default: True] |
dim | Figure size [Tuple of two floats (width, height) in inches][default: (6, 4)] |
theme | Change background theme. If theme set to dark, the dark background will be produced instead of white [string][default:'None'] |
Returns:
PCA biplot 2D and 3D image (biplot_2d.png and biplot_3d.png will be saved in same directory)
latest update v2.0.1
bioinfokit.visuz.cluster.tsneplot(score, colorlist, axlabelfontsize, axlabelfontname, figtype, r, show, markerdot, dotsize, valphadot, colordot, dim, figname, legendpos, legendanchor, theme)
| Parameters | Description |
|---|---|
score | t-SNE component embeddings (obtained from TSNE().fit_transfrom() function in sklearn.manifold) |
colorlist | list of the categories to assign the color [list][default:None] |
axlabelfontsize | Font size for axis labels [float][default: 9] |
axlabelfontname | Font name for axis labels [string][default: 'Arial'] |
figtype | Format of figure to save. Supported format are eps, pdf, pgf, png, ps, raw, rgba, svg, svgz [string][default:'png'] |
r | Figure resolution in dpi [int][default: 300] |
show | Show the figure on console instead of saving in current folder [True or False][default:False] |
markerdot | Shape of the dot on plot. See more options at https://matplotlib.org/3.1.1/api/markers_api.html [string][default: "o"] |
dotsize | The size of the dots in the plot [float][default: 6] |
valphadot | Transparency of dots on plot [float (between 0 and 1)][default: 1] |
colordot | Color of dots on plot [string or list ][default:"#4a4e4d"] |
legendpos | position of the legend on plot. For more options see loc parameter at https://matplotlib.org/3.1.1/api/_as_gen/matplotlib.pyplot.legend.html [string ][default:"best"] |
legendanchor | position of the legend outside of the plot. For more options see bbox_to_anchor parameter at https://matplotlib.org/3.1.1/api/_as_gen/matplotlib.pyplot.legend.html [list][default:None] |
dim | Figure size [Tuple of two floats (width, height) in inches][default: (6, 4)] |
figname | name of figure [string ][default:"tsne_2d"] |
theme | Change background theme. If theme set to dark, the dark background will be produced instead of white [string][default:'None'] |
Returns:
t-SNE 2D image (tsne_2d.png will be saved in same directory)
latest update v0.8.9
Normalize raw gene expression counts into Reads per million mapped reads (RPM) or Counts per million mapped reads (CPM)
bioinfokit.analys.norm.cpm(df)
| Parameters | Description |
|---|---|
df | Pandas dataframe containing raw gene expression values. Genes with missing expression values (NA) will be dropped. |
Returns:
RPM or CPM normalized Pandas dataframe as class attributes (cpm_norm)
latest update v0.9
Normalize raw gene expression counts into Reads per kilo base per million mapped reads (RPKM) or Fragments per kilo base per million mapped reads (FPKM)
bioinfokit.analys.norm.rpkm(df, gl)
| Parameters | Description |
|---|---|
df | Pandas dataframe containing raw gene expression values. Genes with missing expression or gene length values (NA) will be dropped. |
gl | Name of a column having gene length in bp [string][default: None] |
Returns:
RPKM or FPKM normalized Pandas dataframe as class attributes (rpkm_norm)
latest update v0.9.1
Normalize raw gene expression counts into Transcript per million (TPM)
bioinfokit.analys.norm.tpm(df, gl)
| Parameters | Description |
|---|---|
df | Pandas dataframe containing raw gene expression values. Genes with missing expression or gene length values (NA) will be dropped. |
gl | Name of a column having gene length in bp [string][default: None] |
Returns:
TPM normalized Pandas dataframe as class attributes (tpm_norm)
latest update v2.0.1
bioinfokit.visuz.marker.mhat(df, chr, pv, log_scale, color, dim, r, ar, gwas_sign_line, gwasp, dotsize, markeridcol, markernames, gfont, valpha, show, figtype, axxlabel, axylabel, axlabelfontsize, ylm, gstyle, figname, theme)
| Parameters | Description |
|---|---|
df | Pandas dataframe object with atleast SNP, chromosome, and P-values columns |
chr | Name of a column having chromosome numbers [string][default:None] |
pv | Name of a column having P-values. Must be numeric column [string][default:None] |
log_scale | Change the values provided in pv column to minus log10 scale. If set to False, the original values in pv will be used. This is useful in case of Fst values. [Boolean (True or False)][default:True] |
color | List the name of the colors to be plotted. It can accept two alternate colors or the number colors equal to chromosome number. If nothing (None) provided, it will randomly assign the color to each chromosome [list][default:None] |
gwas_sign_line | Plot statistical significant threshold line defined by option gwasp [Boolean (True or False)][default: False] |
gwasp | Statistical significant threshold to identify significant SNPs [float][default: 5E-08] |
dotsize | The size of the dots in the plot [float][default: 8] |
markeridcol | Name of a column having SNPs. This is necessary for plotting SNP names on the plot [string][default: None] |
markernames | The list of the SNPs to display on the plot. These SNP should be present in SNP column. Additionally, it also accepts the dict of SNPs and its associated gene name. If this option set to True, it will label all SNPs with P-value significant score defined by gwasp [string, list, Tuple, dict][default: True] |
gfont | Font size for SNP names to display on the plot [float][default: 8]. gfont not compatible with gstyle=2. |
valpha | Transparency of points on plot [float (between 0 and 1)][default: 1.0] |
dim | Figure size [Tuple of two floats (width, height) in inches][default: (6, 4)] |
r | Figure resolution in dpi [int][default: 300] |
ar | Rotation of X-axis labels [float][default: 90] |
figtype | Format of figure to save. Supported format are eps, pdf, pgf, png, ps, raw, rgba, svg, svgz [string][default:'png'] |
show | Show the figure on console instead of saving in current folder [Boolean (True or False)][default:False] |
axxlabel | Label for X-axis. If you provide this option, default label will be replaced [string][default: None] |
axylabel | Label for Y-axis. If you provide this option, default label will be replaced [string][default: None] |
axlabelfontsize | Font size for axis labels [float][default: 9] |
ylm | Range of ticks to plot on Y-axis [float Tuple (bottom, top, interval)][default: None] |
gstyle | Style of the text for markernames. 1 for default text and 2 for box text [int][default: 1] |
figname | name of figure [string][default:"manhattan"] |
theme | Change background theme. If theme set to dark, the dark background will be produced instead of white [string][default:'None'] |
Returns:
Manhattan plot image in same directory (Manhattan.png)
latest update v0.9.3
Assign genetic features and function to the variants in VCF file
bioinfokit.analys.marker.vcf_anot(file, id, gff_file, anot_attr)
| Parameters | Description |
|---|---|
file | VCF file |
id | chromosome id column in VCF file [string][default='#CHROM'] |
gff_file | GFF3 genome annotation file |
anot_attr | Gene function tag in attributes field of GFF3 file |
Returns:
Tab-delimited text file with annotation (annotated text file will be saved in same directory)
latest update v0.9.4
Concatenate multiple VCF files into single VCF file (for example, VCF files for each chromosome)
bioinfokit.analys.marker.concatvcf(file)
| Parameters | Description |
|---|---|
file | Multiple vcf files separated by comma |
Returns:
Concatenated VCF file (concat_vcf.vcf)
bioinfokit.analys.marker.splitvcf(file)
Split single VCF file containing variants for all chromosomes into individual file containing variants for each chromosome
| Parameters | Description |
|---|---|
file | VCF file to split |
id | chromosome id column in VCF file [string][default='#CHROM'] |
Returns:
VCF files for each chromosome
latest update v0.9.7
bioinfokit.analys.fastq.sra_bd(file, t, other_opts)
FASTQ files will be downloaded using fasterq-dump. Make sure you have the latest version of the NCBI SRA toolkit
(version 2.10.8) is installed and binaries are added to the system path
| Parameters | Description |
|---|---|
file | List of SRA accessions for batch download. All accession must be separated by a newline in the file. |
t | Number of threads for parallel run [int][default=4] |
other_opts | Provide other relevant options for fasterq-dump [str][default=None] Provide the options as a space-separated string. You can get a detailed option for fasterq-dump using the -help option. |
Returns:
FASTQ files for each SRA accession in the current directory unless specified by other_opts
Description and working example
bioinfokit.analys.format.fq_qual_var(file)
| Parameters | Description |
|---|---|
file | FASTQ file to detect quality format [deafult: None] |
Returns:
Quality format encoding name for FASTQ file (Supports only Sanger, Illumina 1.8+ and Illumina 1.3/1.4)
latest update v0.9.7
bioinfokit.analys.fastq.seqcov(file, gs)
| Parameters | Description |
|---|---|
file | FASTQ file |
gs | Genome size in Mbp |
Returns:
Sequencing coverage of the given FASTQ file
Description and Working example
latest update v2.0.6
bioinfokit.analys.Fasta.split_seq(seq, seq_size, seq_overlap, any_cond, outfmt)
| Parameters | Description |
|---|---|
seq | Input sequence [string] |
seq_size | subsequence size [int][default: 3] |
seq_overlap | Split the sequence in overlap mode [bool][default: True] |
any_cond | Split sequence based on a condition. Note yet defined. |
outfmt | Output format for the subsequences. If parameter set to 'fasta', the file will be saved in same folder with name output_chunks.fasta ['list' or 'fasta'][default: 'list'] |
Returns:
Subsequences in list or fasta file (output_chunks.fasta) format
Description and Working example
latest update v2.1.1
bioinfokit.analys.Fasta.rev_com(sequence)
| Parameters | Description |
|---|---|
seq | DNA sequence to perform reverse complement |
file | DNA sequence in a fasta file |
Returns:
Reverse complement of original DNA sequence
bioinfokit.analys.format
| Function | Parameters | Description |
|---|---|---|
bioinfokit.analys.format.fqtofa(file) | FASTQ file | Convert FASTQ file into FASTA format |
bioinfokit.analys.format.hmmtocsv(file) | HMM file | Convert HMM text output (from HMMER tool) to CSV format |
bioinfokit.analys.format.tabtocsv(file) | TAB file | Convert TAB file to CSV format |
bioinfokit.analys.format.csvtotab(file) | CSV file | Convert CSV file to TAB format |
Returns:
Output will be saved in same directory
latest update v1.0.1
bioinfokit.analys.gff.gff_to_gtf(file, trn_feature_name)
| Parameters | Description |
|---|---|
file | GFF3 genome annotation file |
trn_feature_name | Name of the feature (column 3 of GFF3 file) of RNA transcripts if other than 'mRNA' or 'transcript' |
Returns:
GTF format genome annotation file (file.gtf will be saved in same directory)
latest update v2.0.4
| Function | Parameters | Description |
|---|---|---|
bioinfokit.analys.Fasta.fasta_reader(file) | FASTA file | FASTA file reader |
bioinfokit.analys.fastq.fastq_reader(file) | FASTQ file | FASTQ file reader |
bioinfokit.analys.marker.vcfreader(file) | VCF file | VCF file reader |
Returns:
File generator object (can be iterated only once) that can be parsed for the record
Description and working example
latest update v2.0.4
bioinfokit.analys.Fasta.ext_subseq(file, id, st, end, strand)
Extract the subsequence of specified region from FASTA file. If the target subsequence region is on minus strand. the reverse complementary of subsequence will be printed.
| Parameters | Description |
|---|---|
file | FASTA file [file] |
id | The ID of sequence from FASTA file to extract the subsequence [string] |
st | Start integer coordinate of subsequnece [int] |
end | End integer coordinate of subsequnece [int] |
strand | Strand of the subsequence ['plus' or 'minus'][default: 'plus'] |
Returns:
Subsequence to stdout
latest update v2.1.3
bioinfokit.analys.Fasta.extract_seq(file, id)
Extract the sequences from FASTA file based on the list of sequence IDs provided from other file
| Parameters | Description |
|---|---|
file | FASTA file [file] |
id | List of sequence IDs separated by new line. This file can also contain the ID, start and end coordinates separated by TAB [file] |
Returns:
Sequences extracted from FASTA file based on the given IDs provided in id file. Output FASTA file will be saved as output.fasta in current working directory.
Description and working example
latest update v2.0.4
bioinfokit.analys.Fasta.split_fasta(file, n, bases_per_line)
Split one big FASTA file into multiple smaller FASTA files
| Parameters | Description |
|---|---|
file | FASTA file [file] |
n | Number of FASTA files to split the big FASTA file [int][default: 2] |
bases_per_line | Number of bases per line for ouput FASTA files [int][default: 60] |
Returns:
Number of smaller FASTA files with prefix output (output_0.fasta, output_1.fasta and so on)
latest update v2.1.2
bioinfokit.analys.Fasta.multi_to_single_line(file)
Convert multi-line FASTA (where sequences are on multi lines) into single-line FASTA (where sequences are in single line)
| Parameters | Description |
|---|---|
file | FASTA file [file] |
Returns:
Single line FASTA (output.fasta). Output FASTA file will be saved as output.fasta in current working directory.
Description and working example
latest update v2.0.5
bioinfokit.analys.HtsAna.merge_featureCount(pattern, gene_column_name)
Merge counts files generated from featureCounts when it runs individually on large samples. The count files must be in same folder and should end with .txt file extension.
| Parameters | Description |
|---|---|
pattern | file name pattern for each count file [default: '*.txt'] |
gene_column_name | gene id column name for feature and meta-features [default: 'Geneid'] |
Returns:
Merge count file (gene_matrix_count.csv) in same folder
latest update v2.0.9
bioinfokit.analys.HtsAna.split_bed(bed)
Split the BED file by chromosome names
| Parameters | Description |
|---|---|
bed | Input BED file [default: None] |
Returns:
BED file for each chromosome (files will be saved in same directory)
latest update v2.1.4
bioinfokit.analys.Fasta.max_min_len(fasta)
Find Max and Min sequence lengths from Fasta
| Parameters | Description |
|---|---|
fasta | Input Fasta file [default: None] |
Returns:
Max and Min sequence lengths from Fasta file
latest update v1.0.0
bioinfokit.analys.genfam.fam_enrich(id_file, species, id_type, stat_sign_test, multi_test_corr, min_map_ids, alpha)
GenFam is a comprehensive classification and enrichment analysis tool for plant genomes. It provides a unique way to characterize the large-scale gene datasets such as those from transcriptome analysis (read GenFam paper for more details)
| Parameters | Description |
|---|---|
id_file | Text file containing the list of gene IDs to analyze using GenFam. IDs must be separated by newline. |
species | Plant species ID for GenFam analysis. All plant species ID provided here |
id_type | Plant species ID type 1: Phytozome locus ID 2: Phytozome transcript ID 3: Phytozome PAC ID |
stat_sign_test | Statistical significance test for enrichment analysis [default=1]. 1: Fisher exact test 2: Hypergeometric distribution 3: Binomial distribution 4: Chi-squared distribution |
multi_test_corr | Multiple testing correction test [default=3]. 1: Bonferroni 2: Bonferroni-Holm 3: Benjamini-Hochberg |
min_map_ids | Minimum number of gene IDs from the user list (id_file) must be mapped to the background database for performing GenFam analysis [default=5] |
alpha | Significance level [float][default: 0.05] |
Returns:
| Attribute | Description |
|---|---|
| df_enrich | Enriched gene families with p < 0.05 |
| genfam_info | GenFam run information |
| Output files | Output figures and files from GenFam analysis genfam_enrich.png: GenFam figure for enriched gene families fam_enrich_out.txt: List of enriched gene families with mapped gene IDs, GO annotation, and detailed statistics fam_all_out.txt: List of all gene families with mapped gene IDs, GO annotation, and detailed statistics |
Description and working example
latest update v1.0.0
bioinfokit.analys.genfam.check_allowed_ids(species)
| Parameters | Description |
|---|---|
species | Plant species ID to check for allowed ID type. All plant species ID provided here |
Returns:
Allowed ID types for GenFam
Description and working example
latest update v2.0.1
bioinfokit.visuz.stat.corr_mat(table, corm, cmap, r, dim, show, figtype, axtickfontsize, axtickfontname, theme)
| Parameters | Description |
|---|---|
table | Dataframe object with numerical variables (columns) to find correlation. Ideally, you should have three or more variables. Dataframe should not have identifier column. |
corm | Correlation method [pearson,kendall,spearman] [default:pearson] |
cmap | Color Palette for heatmap [string][default: 'seismic']. More colormaps are available at https://matplotlib.org/3.1.0/tutorials/colors/colormaps.html |
r | Figure resolution in dpi [int][default: 300]. Not compatible with show= True |
dim | Figure size [Tuple of two floats (width, height) in inches][default: (6, 5)] |
show | Show the figure on console instead of saving in current folder [True or False][default:False] |
figtype | Format of figure to save. Supported format are eps, pdf, pgf, png, ps, raw, rgba, svg, svgz [string][default:'png'] |
axtickfontsize | Font size for axis ticks [float][default: 7] |
axtickfontname | Font name for axis ticks [string][default: 'Arial'] |
theme | Change background theme. If theme set to dark, the dark background will be produced instead of white [string][default:'None'] |
Returns:
Correlation matrix plot image in same directory (corr_mat.png)
latest update v0.8.5
bioinfokit.visuz.stat.bardot(df, colorbar, colordot, bw, dim, r, ar, hbsize, errorbar, dotsize, markerdot, valphabar, valphadot, show, figtype, axxlabel, axylabel, axlabelfontsize, axlabelfontname, ylm, axtickfontsize, axtickfontname, yerrlw, yerrcw)
| Parameters | Description |
|---|---|
df | Pandas dataframe object |
colorbar | Color of bar graph [string or list][default:"#bbcfff"] |
colordot | Color of dots on bar [string or list][default:"#ee8972"] |
bw | Width of bar [float][default: 0.4] |
dim | Figure size [Tuple of two floats (width, height) in inches][default: (6, 4)] |
r | Figure resolution in dpi [int][default: 300] |
ar | Rotation of X-axis labels [float][default: 0] |
hbsize | Horizontal bar size for standard error bars [float][default: 4] |
errorbar | Draw standard error bars [bool (True or False)][default: True] |
dotsize | The size of the dots in the plot [float][default: 6] |
markerdot | Shape of the dot marker. See more options at https://matplotlib.org/3.1.1/api/markers_api.html [string][default: "o"] |
valphabar | Transparency of bars on plot [float (between 0 and 1)][default: 1] |
valphadot | Transparency of dots on plot [float (between 0 and 1)][default: 1] |
figtype | Format of figure to save. Supported format are eps, pdf, pgf, png, ps, raw, rgba, svg, svgz [string][default:'png'] |
show | Show the figure on console instead of saving in current folder [True or False][default:False] |
axxlabel | Label for X-axis. If you provide this option, default label will be replaced [string][default: None] |
axylabel | Label for Y-axis. If you provide this option, default label will be replaced [string][default: None] |
axlabelfontsize | Font size for axis labels [float][default: 9] |
axlabelfontname | Font name for axis labels [string][default: 'Arial'] |
ylm | Range of ticks to plot on Y-axis [float Tuple (bottom, top, interval)][default: None] |
axtickfontsize | Font size for axis ticks [float][default: 9] |
axtickfontname | Font name for axis ticks [string][default: 'Arial'] |
yerrlw | Error bar line width [float][default: None] |
yerrcw | Error bar cap width [float][default: None] |
Returns:
Bar-dot plot image in same directory (bardot.png)
latest update v2.1.0
bioinfokit.analys.stat.ztest(df, x, y, mu, x_std, y_std, alpha, test_type)
| Parameters | Description |
|---|---|
df | Pandas dataframe for appropriate Z-test. One sample: It should have atleast one variable Two sample independent: It should have atleast two variables |
x | column name for x group [string][default: None] |
y | column name for x group [string][default: None] |
mu | Population or known mean for the one sample Z-test [float][default: None] |
x_std | Population standard deviation for x group [float][default: None] |
y_std | Population standard deviation for y group [float][default: None] |
alpha | Significance level for confidence interval (CI). If alpha=0.05, then 95% CI will be calculated [float][default: 0.05] |
test_type | Type of Z-test [int (1,2)][default: None]. 1: One sample Z-test 2: Two sample Z-test |
Returns:
Summary output as class attribute (summary and result)
Description and Working example
latest update v2.1.0
bioinfokit.analys.stat.ttest(df, xfac, res, evar, alpha, test_type, mu)
| Parameters | Description |
|---|---|
df | Pandas dataframe for appropriate t-test. One sample: It should have atleast dependent (res) variable Two sample independent: It should have independent (xfac) and dependent (res) variables Two sample paired: It should have two dependent (res) variables |
xfac | Independent group column name with two levels [string][default: None] |
res | Dependent variable column name [string or list or Tuple][default: None] |
evar | t-test with equal variance [bool (True or False)][default: True] |
alpha | Significance level for confidence interval (CI). If alpha=0.05, then 95% CI will be calculated [float][default: 0.05] |
test_type | Type of t-test [int (1,2,3)][default: None]. 1: One sample t-test 2: Two sample independent t-test 3: Two sample paired t-test |
mu | Population or known mean for the one sample t-test [float][default: None] |
Returns:
Summary output as class attribute (summary and result)
Description and Working example
latest update v0.9.5
bioinfokit.analys.stat.chisq(df, p)
| Parameters | Description |
|---|---|
df | Pandas dataframe. It should be one or two-dimensional contingency table. |
p | Theoretical expected probabilities for each group. It must be non-negative and sum to 1. If p is provide Goodness of Fit test will be performed [list or Tuple][default: None] |
Returns:
Summary and expected counts as class attributes (summary and expected_df)
bioinfokit.visuz.stat.lin_reg(df, x, y)
| Parameters | Description |
|---|---|
df | Pandas dataframe object |
x | Name of column having independent X variables [list][default:None] |
y | Name of column having dependent Y variables [list][default:None] |
Returns:
Regression analysis summary
latest update v2.0.1
bioinfokit.visuz.stat.regplot(df, x, y, yhat, dim, colordot, colorline, r, ar, dotsize, markerdot, linewidth, valphaline, valphadot, show, figtype, axxlabel, axylabel, axlabelfontsize, axlabelfontname, xlm, ylm, axtickfontsize, axtickfontname, theme)
| Parameters | Description |
|---|---|
df | Pandas dataframe object |
x | Name of column having independent X variables [string][default:None] |
y | Name of column having dependent Y variables [string][default:None] |
yhat | Name of column having predicted response of Y variable (y_hat) from regression [string][default:None] |
dim | Figure size [Tuple of two floats (width, height) in inches][default: (6, 4)] |
r | Figure resolution in dpi [int][default: 300] |
ar | Rotation of X-axis labels [float][default: 0] |
dotsize | The size of the dots in the plot [float][default: 6] |
markerdot | Shape of the dot marker. See more options at https://matplotlib.org/3.1.1/api/markers_api.html [string][default: "o"] |
valphaline | Transparency of regression line on plot [float (between 0 and 1)][default: 1] |
valphadot | Transparency of dots on plot [float (between 0 and 1)][default: 1] |
linewidth | Width of regression line [float][default: 1] |
figtype | Format of figure to save. Supported format are eps, pdf, pgf, png, ps, raw, rgba, svg, svgz [string][default:'png'] |
show | Show the figure on console instead of saving in current folder [True or False][default:False] |
axxlabel | Label for X-axis. If you provide this option, default label will be replaced [string][default: None] |
axylabel | Label for Y-axis. If you provide this option, default label will be replaced [string][default: None] |
axlabelfontsize | Font size for axis labels [float][default: 9] |
axlabelfontname | Font name for axis labels [string][default: 'Arial'] |
xlm | Range of ticks to plot on X-axis [float Tuple (bottom, top, interval)][default: None] |
ylm | Range of ticks to plot on Y-axis [float Tuple (bottom, top, interval)][default: None] |
axtickfontsize | Font size for axis ticks [float][default: 9] |
axtickfontname | Font name for axis ticks [string][default: 'Arial'] |
theme | Change background theme. If theme set to dark, the dark background will be produced instead of white [string][default:'None'] |
Returns:
Regression plot image in same directory (reg_plot.png)
latest update v1.0.3
bioinfokit.analys.stat.tukey_hsd(df, res_var, xfac_var, anova_model, phalpha, ss_typ)
It performs multiple pairwise comparisons of treatment groups using Tukey's HSD (Honestly Significant Difference) test to check if group means are significantly different from each other. It uses the Tukey-Kramer approach if the sample sizes are unequal among the groups.
| Parameters | Description |
|---|---|
df | Pandas dataframe with the variables mentioned in the res_var, xfac_var and anova_model options. It should not have missing data. The missing data will be omitted. |
res_var | Name of a column having response variable [string][default: None] |
xfac_var | Name of a column having factor or group for pairwise comparison [string][default: None] |
anova_model | ANOVA model (calculated using statsmodels ols function) [string][default: None] |
phalpha | Significance level [float][default: 0.05] |
ss_typ | Type of sum of square to perform ANOVA [int][default: 2] |
Returns:
| Attribute | Description |
|---|---|
tukey_summary | Pairwise comparisons for main and interaction effects by Tukey HSD test |
Description and Working example
latest update v1.0.3
bioinfokit.analys.stat.bartlett(df, xfac_var, res_var)
It performs Bartlett's test to check the homogeneity of variances among the treatment groups. It accepts the input table in a stacked format. More details https://docs.scipy.org/doc/scipy-0.14.0/reference/generated/scipy.stats.bartlett.html
| Parameters | Description |
|---|---|
df | Pandas dataframe containing response (res_var) and independent variables (xfac_var) in a stacked format. It should not have missing data. The missing data will be omitted. |
res_var | Name of a column having response variable [string][default: None] |
xfac_var | Name of a column having treatment groups (independent variables) [string or list][default: None] |
Returns:
| Attribute | Description |
|---|---|
bartlett_summary | Pandas dataframe containing Bartlett's test statistics, degree of freedom, and p value |
Description and Working example
latest update v1.0.3
bioinfokit.analys.stat.levene(df, xfac_var, res_var)
It performs Levene's test to check the homogeneity of variances among the treatment groups. It accepts the input table in a stacked format. More details https://docs.scipy.org/doc/scipy-0.14.0/reference/generated/scipy.stats.levene.html
| Parameters | Description |
|---|---|
df | Pandas dataframe containing response (res_var) and independent variables (xfac_var) in a stacked format. It should not have missing data. The missing data will be omitted. |
res_var | Name of a column having response variable [string][default: None] |
xfac_var | Name of a column having treatment groups (independent variables) [string or list][default: None] |
center | Choice for the Levene's test [string (median, mean, trimmed)] [default: median] median: Brown-Forsythe Levene-type test mean: original Levene's test trimmed: Brown-Forsythe Levene-type test |
Returns:
| Attribute | Description |
|---|---|
levene_summary | Pandas dataframe containing Levene's test statistics, degree of freedom, and p value |
Description and Working example
latest update v2.0.1
bioinfokit.visuz.stat.roc(fpr, tpr, c_line_style, c_line_color, c_line_width, diag_line, diag_line_style, diag_line_width, diag_line_color, auc, shade_auc, shade_auc_color, axxlabel, axylabel, axtickfontsize, axtickfontname, axlabelfontsize, axlabelfontname, plotlegend, legendpos, legendanchor, legendcols, legendfontsize, legendlabelframe, legend_columnspacing, dim, show, figtype, figname, r, ylm, theme)
Receiver operating characteristic (ROC) curve for visualizing classification performance
| Parameters | Description |
|---|---|
fpr | Increasing false positive rates obtained from sklearn.metrics.roc_curve [list][default:None] |
tpr | Increasing true positive rates obtained from sklearn.metrics.roc_curve [list][default:None] |
c_line_style | Line style for ROC curve [string][default:'-'] |
c_line_color | Line color for ROC curve [string][default:'#f05f21'] |
c_line_width | Line width for ROC curve [float][default:1] |
diag_line | Plot reference line [True or False][default: True] |
diag_line_style | Line style for reference line [string][default:'--'] |
diag_line_width | Line width for reference line [float][default:1] |
diag_line_color | Line color for reference line [string][default:'b'] |
auc | Area under ROC. It can be obtained from sklearn.metrics.roc_auc_score [float][default: None] |
shade_auc | Shade are for AUC [True or False][default: False] |
shade_auc_color | Shade color for AUC [string][default: '#f48d60'] |
axxlabel | Label for X-axis [string][default: 'False Positive Rate (1 - Specificity)'] |
axylabel | Label for Y-axis [string][default: 'True Positive Rate (Sensitivity)'] |
axtickfontsize | Font size for axis ticks [float][default: 9] |
axtickfontname | Font name for axis ticks [string][default: 'Arial'] |
axlabelfontsize | Font size for axis labels [float][default: 9] |
axlabelfontname | Font name for axis labels [string][default: 'Arial'] |
plotlegend | plot legend [True or False][default:True] |
legendpos | position of the legend on plot. For more options see loc parameter at https://matplotlib.org/3.1.1/api/_as_gen/matplotlib.pyplot.legend.html [string ][default:'lower right'] |
legendanchor | position of the legend outside of the plot. For more options see bbox_to_anchor parameter at https://matplotlib.org/3.1.1/api/_as_gen/matplotlib.pyplot.legend.html [list][default:None] |
legendcols | Number of columns for legends [int][default: 1] |
legendfontsize | Font size for the legends [float][default:8] |
legendlabelframe | Box frame for the legend [True or False][default: False] |
legend_columnspacing | Spacing between the legends [float][default: None] |
dim | Figure size [Tuple of two floats (width, height) in inches][default: (5, 4)] |
show | Show the figure on console instead of saving in current folder [True or False][default:False] |
figtype | Format of figure to save. Supported format are eps, pdf, pgf, png, ps, raw, rgba, svg, svgz [string][default:'png'] |
figname | name of figure [string ][default:'roc'] |
r | Figure resolution in dpi [int][default: 300]. Not compatible with show= True |
ylm | Range of ticks to plot on Y-axis [float (bottom, top, interval)][default: None] |
theme | Change background theme. If theme set to dark, the dark background will be produced instead of white [string][default:'None'] |
Returns:
ROC plot image in same directory (roc.png) Working example
Calculate Root Mean Square Error (RMSE), Mean squared error (MSE), Mean absolute error (MAE), and Mean absolute percent error (MAPE) from regression fit
latest update v1.0.8
bioinfokit.analys.stat.reg_metric(y, yhat, resid)
| Parameters | Description |
|---|---|
y | Original values for dependent variable [numpy array] [default: None] |
yhat | Predicted values from regression [numpy array] [default: None] |
resid | Regression residuals [numpy array][default: None] |
Returns:
Pandas dataframe with values for RMSE, MSE, MAE, and MAPE
Venn Diagram
bioinfokit.visuz.venn(vennset, venncolor, vennalpha, vennlabel)
| Parameters | Description |
|---|---|
vennset | Venn dataset for 3 and 2-way venn. Data should be in the format of (100,010,110,001,101,011,111) for 3-way venn and 2-way venn (10, 01, 11) [default: (1,1,1,1,1,1,1)] |
venncolor | Color Palette for Venn [color code][default: ('#00909e', '#f67280', '#ff971d')] |
vennalpha | Transparency of Venn [float (0 to 1)][default: 0.5] |
vennlabel | Labels to Venn [string][default: ('A', 'B', 'C')] |
Returns:
Venn plot (venn3.png, venn2.png)
Last updated: November 20, 2021
FAQs
Bioinformatics data analysis and visualization toolkit
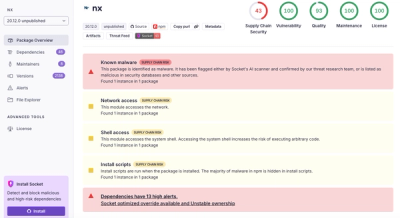
We found that bioinfokit demonstrated a healthy version release cadence and project activity because the last version was released less than a year ago. It has 1 open source maintainer collaborating on the project.
Did you know?

Socket for GitHub automatically highlights issues in each pull request and monitors the health of all your open source dependencies. Discover the contents of your packages and block harmful activity before you install or update your dependencies.

Security News
/Research
Malicious npm package impersonates Nodemailer and drains wallets by hijacking crypto transactions across multiple blockchains.

Security News
This episode explores the hard problem of reachability analysis, from static analysis limits to handling dynamic languages and massive dependency trees.
Security News
/Research
Malicious Nx npm versions stole secrets and wallet info using AI CLI tools; Socket’s AI scanner detected the supply chain attack and flagged the malware.