Research
/Security News
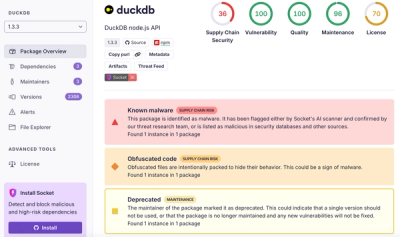
DuckDB npm Account Compromised in Continuing Supply Chain Attack
Ongoing npm supply chain attack spreads to DuckDB: multiple packages compromised with the same wallet-drainer malware.
MCP (Model Context Protocol) server for Boltz-2 protein structure prediction model.
This MCP server provides tools for protein structure prediction using the state-of-the-art Boltz-2 model. It enables you to:
boltz_predict_structure - Main prediction tool with full configuration optionsboltz_predict_from_sequences - Simple sequence-to-structure predictionboltz_check_status - Check Boltz installation statusboltz_get_examples - Get example configurations for different prediction typesresource://boltz-usage-guide - Comprehensive usage guide and documentationBefore installing the Python packages, you need to install system dependencies required for building scientific Python packages:
Ubuntu/Debian:
sudo apt update
sudo apt install -y cmake build-essential gfortran
CentOS/RHEL/Fedora:
# For CentOS/RHEL
sudo yum install cmake gcc-gfortran gcc-c++ make
# For Fedora
sudo dnf install cmake gcc-gfortran gcc-c++ make
macOS:
# Install Xcode Command Line Tools if not already installed
xcode-select --install
# Install gfortran (via Homebrew)
brew install gcc cmake
Note: These system dependencies are required because some Python packages (like SciPy) need to be compiled from source when using uv, and they require:
cmake: For building dm-tree and other native extensionsgfortran: For building SciPy from sourcebuild-essential/gcc-c++: For general C/C++ compilationInstall the Boltz-2 model:
pip install boltz
Install this MCP server:
cd boltz-mcp
uv pip install -e .
Boltz-2 works best with GPU acceleration. Ensure you have:
You do not have to copy anything, just use uvx and it will install and run the MCP. Note: the download can take a lot because of evil CUDA dependencies.
uvx --python 3.12 boltz-mcp stdio
Clone the repo and run with uv
gh repo clone longevity-genie/boltz-mcp
cd boltz-mcp
# or
uv run boltz-mcp
If you want to inspect the methods provided by the MCP server, use npx (you may need to install nodejs and npm):
For STDIO mode:
npx @modelcontextprotocol/inspector --config mcp-config-local.json --server boltz-mcp
For local stdio mode:
npx @modelcontextprotocol/inspector --config mcp-config-local.json --server boltz-mcp
You can also run the inspector manually and configure it through the interface:
npx @modelcontextprotocol/inspector
After that you can explore the tools and resources with MCP Inspector at http://127.0.0.1:6274 (note, if you run inspector several times it can change port, also use the url that includes proxy token when possible)
Note: Using the MCP Inspector is optional. Most MCP clients (like Cursor, Windsurf, Claude Desktop, etc.) will automatically display the available tools from this server once configured. However, the Inspector can be useful for detailed testing and exploration.
If you choose to use the Inspector via npx, ensure you have Node.js and npm installed. Using nvm (Node Version Manager) is recommended for managing Node.js versions.
# Using the boltz_predict_from_sequences tool
{
"sequences": {
"sequences": {
"protein1": "MKTVRQERLKSIVRILERSKEPVSGAQLAEELSVSRQVIVQDIAYLRSLGYNIVATPRGYVLAGG"
}
},
"output_dir": "./predictions",
"model": "boltz2"
}
# Using the boltz_predict_structure tool
{
"input_data": {
"sequences": {
"chain_A": "MKTVRQERLKSIVRILERSKEPVSGAQLAEELSVSRQVIVQDIAYLRSLGYNIVATPRGYVLAGG",
"chain_B": "ARNDCEQGHILKMFPSTWYV"
}
},
"model": "boltz2",
"devices": 1,
"accelerator": "gpu",
"sampling_steps": 200,
"diffusion_samples": 1,
"output_format": "mmcif"
}
{
"input_data": {
"sequences": ["protein.fasta"],
"job_name": "my_prediction",
"pdb_id": "1ABC", # Template structure
"msa": ["custom.a3m"], # Custom MSA
"ligands": ["ligand.sdf"] # Ligands
},
"model": "boltz2",
"devices": 2,
"recycling_steps": 5,
"sampling_steps": 300,
"use_msa_server": true,
"override": false
}
The server returns:
Output files include:
model: "boltz1" or "boltz2" (default: "boltz2")devices: Number of GPU devices (default: 1)accelerator: "gpu", "cpu", or "tpu" (default: "gpu")recycling_steps: Number of recycling iterations (default: 3)sampling_steps: Diffusion sampling steps (default: 200)diffusion_samples: Number of samples to generate (default: 1)output_format: "pdb" or "mmcif" (default: "mmcif")output_dir: Where to save resultsuse_msa_server: Use online MSA generationoverride: Override existing predictionsstep_scale: Control sampling temperatureThe server handles various error conditions:
Check the boltz_check_status tool to verify installation.
The Boltz-2 developers have unfortunately pinned minor versions of several critical libraries in their dependency specifications, which can lead to significant compatibility issues:
Workarounds:
These issues stem from upstream dependency management decisions in the Boltz-2 package and are beyond the scope of this MCP server.
boltz-mcp/
├── src/
│ └── boltz_mcp/
│ ├── __init__.py
│ └── server.py
├── pyproject.toml
└── README.md
This project follows the same license as the original Boltz-2 model.
Contributions are welcome! Please submit issues and pull requests on the project repository.
For issues related to:
FAQs
MCP for Boltz-2 model inference
We found that boltz-mcp demonstrated a healthy version release cadence and project activity because the last version was released less than a year ago. It has 1 open source maintainer collaborating on the project.
Did you know?

Socket for GitHub automatically highlights issues in each pull request and monitors the health of all your open source dependencies. Discover the contents of your packages and block harmful activity before you install or update your dependencies.

Research
/Security News
Ongoing npm supply chain attack spreads to DuckDB: multiple packages compromised with the same wallet-drainer malware.

Security News
The MCP Steering Committee has launched the official MCP Registry in preview, a central hub for discovering and publishing MCP servers.
Product
Socket’s new Pull Request Stories give security teams clear visibility into dependency risks and outcomes across scanned pull requests.