|PyPI| |Downloads| |CI| |Docs| |Codecov| |Discourse|
CellRank 2: Unified fate mapping in multiview single-cell data
.. image:: docs/_static/img/light_mode_overview.png#gh-light-mode-only
:width: 600px
:align: center
:class: only-light
.. image:: docs/_static/img/dark_mode_overview.png#gh-dark-mode-only
:width: 600px
:align: center
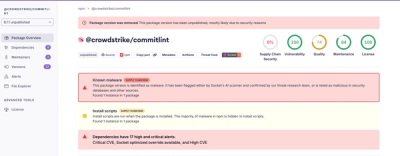
CellRank is a modular framework to study cellular dynamics based on Markov state modeling of
multi-view single-cell data. See our documentation, and the CellRank 1 and CellRank 2 manuscript_ to learn more.
.. important::
Please refer to our citation guide <https://github.com/theislab/cellrank/blob/main/docs/about/cite.rst>_ to cite our software correctly.
CellRank scales to large cell numbers, is fully compatible with the scverse_ ecosystem, and easy to use.
In the backend, it is powered by pyGPCCA_ (Reuter et al. (2018)). Feel
free to open an issue if you encounter a bug, need our help or just want to make a comment/suggestion.
CellRank's key applications
- Estimate differentiation direction based on a varied number of biological priors, including RNA velocity
(
La Manno et al. (2018), Bergen et al. (2020)), any pseudotime or developmental potential,
experimental time points, metabolic labels, and more.
- Compute initial, terminal and intermediate macrostates.
- Infer fate probabilities and driver genes.
- Visualize and cluster gene expression trends.
- ... and much more, check out our
documentation_.
.. |PyPI| image:: https://img.shields.io/pypi/v/cellrank.svg
:target: https://pypi.org/project/cellrank
:alt: PyPI
.. |Downloads| image:: https://static.pepy.tech/badge/cellrank
:target: https://pepy.tech/project/cellrank
:alt: Downloads
.. |Discourse| image:: https://img.shields.io/discourse/posts?color=yellow&logo=discourse&server=https%3A%2F%2Fdiscourse.scverse.org
:target: https://discourse.scverse.org/c/ecosystem/cellrank/
:alt: Discourse
.. |CI| image:: https://img.shields.io/github/actions/workflow/status/theislab/cellrank/test.yml?branch=main
:target: https://github.com/theislab/cellrank/actions
:alt: CI
.. |Docs| image:: https://img.shields.io/readthedocs/cellrank
:target: https://cellrank.readthedocs.io/
:alt: Documentation
.. |Codecov| image:: https://codecov.io/gh/theislab/cellrank/branch/main/graph/badge.svg
:target: https://codecov.io/gh/theislab/cellrank
:alt: Coverage
.. _La Manno et al. (2018): https://doi.org/10.1038/s41586-018-0414-6
.. _Bergen et al. (2020): https://doi.org/10.1038/s41587-020-0591-3
.. _Reuter et al. (2018): https://doi.org/10.1021/acs.jctc.8b00079
.. _scverse: https://scverse.org/
.. _pyGPCCA: https://github.com/msmdev/pyGPCCA
.. _CellRank 1: https://www.nature.com/articles/s41592-021-01346-6
.. _CellRank 2 manuscript: https://doi.org/10.1038/s41592-024-02303-9
.. _documentation: https://cellrank.org
.. _issue: https://github.com/theislab/cellrank/issues/new/choose