
Security News
curl Shuts Down Bug Bounty Program After Flood of AI Slop Reports
A surge of AI-generated vulnerability reports has pushed open source maintainers to rethink bug bounties and tighten security disclosure processes.
dnaio
Advanced tools
.. image:: https://zenodo.org/badge/DOI/10.5281/zenodo.10548864.svg :target: https://doi.org/10.5281/zenodo.10548864
.. image:: https://github.com/marcelm/dnaio/workflows/CI/badge.svg :alt: GitHub Actions badge
.. image:: https://img.shields.io/pypi/v/dnaio.svg?branch=main :target: https://pypi.python.org/pypi/dnaio :alt: PyPI badge
.. image:: https://codecov.io/gh/marcelm/dnaio/branch/master/graph/badge.svg :target: https://codecov.io/gh/marcelm/dnaio :alt: Codecov badge
dnaio is a Python 3 library for very efficient parsing and writing of FASTQ and also FASTA files.
Since dnaio version 1.1.0, support for efficiently parsing uBAM files has been implemented.
This allows reading ONT files from the dorado <https://github.com/nanoporetech/dorado>_
basecaller directly.
The code was previously part of the
Cutadapt <https://cutadapt.readthedocs.io/>_ tool and has been improved significantly since it has been split out.
The main interface is the dnaio.open <https://dnaio.readthedocs.io/en/latest/api.html>_ function::
import dnaio
with dnaio.open("reads.fastq.gz") as f:
bp = 0
for record in f:
bp += len(record)
print(f"The input file contains {bp/1E6:.1f} Mbp")
For more, see the tutorial <https://dnaio.readthedocs.io/en/latest/tutorial.html>_ and
API documentation <https://dnaio.readthedocs.io/en/latest/api.html>_.
Using pip::
pip install dnaio zstandard
zstandard can be omitted if support for Zstandard (.zst) files is not required.
.gz, .bz2, .xz and .zst are detected automatically)+) are supportedDocumentation <https://dnaio.readthedocs.io/>_Source code <https://github.com/marcelm/dnaio/>_Report an issue <https://github.com/marcelm/dnaio/issues>_Project page on PyPI <https://pypi.python.org/pypi/dnaio/>_FAQs
Read and write FASTA and FASTQ files efficiently
We found that dnaio demonstrated a healthy version release cadence and project activity because the last version was released less than a year ago. It has 2 open source maintainers collaborating on the project.
Did you know?

Socket for GitHub automatically highlights issues in each pull request and monitors the health of all your open source dependencies. Discover the contents of your packages and block harmful activity before you install or update your dependencies.

Security News
A surge of AI-generated vulnerability reports has pushed open source maintainers to rethink bug bounties and tighten security disclosure processes.
Product
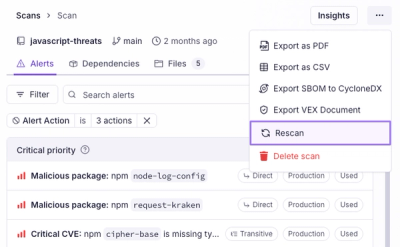
Scan results now load faster and remain consistent over time, with stable URLs and on-demand rescans for fresh security data.
Product
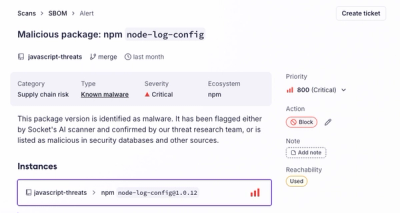
Socket's new Alert Details page is designed to surface more context, with a clearer layout, reachability dependency chains, and structured review.