Security News
Crates.io Users Targeted by Phishing Emails
The Rust Security Response WG is warning of phishing emails from rustfoundation.dev targeting crates.io users.
|pypi| |actions| |codecov|
Reportable clinic events, reference ranges, grading
Normal data is kept in model NormalData and grading data in GradingData.
The two tables are populated by the post_migrate signal post_migrate_load_reference_ranges.
The post-migrate signal goes through all apps looking for reportables.py and loads according
to the attributes in the module.
A reportables.py might look like this:
.. code-block:: python
from edc_reportable.data import africa, daids_july_2017
collection_name = "meta"
normal_data = africa.normal_data
grading_data = {}
grading_data.update(**daids_july_2017.dummies)
grading_data.update(**daids_july_2017.chemistries)
grading_data.update(**daids_july_2017.hematology)
reportable_grades = [3, 4]
reportable_grades_exceptions = {}
These attributes in reportables.py are required:
When the post-migrate signal finds a module it calls load_reference_ranges:
.. code-block:: python
load_reference_ranges(
reportables_module.collection_name,
normal_data=reportables_module.normal_data,
grading_data=reportables_module.grading_data,
reportable_grades=reportables_module.reportable_grades,
reportable_grades_exceptions=reportables_module.reportable_grades_exceptions,
)
A normal reference is declared like this:
.. code-block:: python
normal_data = {
"albumin": [
Formula(
"3.5<=x<=5.0",
units=GRAMS_PER_DECILITER,
gender=[MALE, FEMALE],
**adult_age_options,
),
],
...
}
Add as many normal references in the dictionary as you like, just ensure the lower and upper boundaries don't overlap.
Note: If the lower and upper values of a normal reference overlap
with another normal reference in the same group, a BoundaryOverlap
exception will be raised when the value is evaluated.
Catch this in your tests.
See edc_reportable.data.normal_data for a complete example.
A grading reference is declared like this:
.. code-block:: python
from edc_constants.constants import FEMALE, MALE
from ...adult_age_options import adult_age_options
from ...constants import HIGH_VALUE
from ...units import IU_LITER
grading_data = {
amylase=[
Formula(
"1.1*ULN<=x<1.5*ULN",
grade=1,
units=IU_LITER,
gender=[MALE, FEMALE],
**adult_age_options,
),
Formula(
"1.5*ULN<=x<3.0*ULN",
grade=2,
units=IU_LITER,
gender=[MALE, FEMALE],
**adult_age_options,
),
Formula(
"3.0*ULN<=x<5.0*ULN",
grade=3,
units=IU_LITER,
gender=[MALE, FEMALE],
**adult_age_options,
),
Formula(
f"5.0*ULN<=x<{HIGH_VALUE}*ULN",
grade=4,
units=IU_LITER,
gender=[MALE, FEMALE],
**adult_age_options,
),
],
...
}
Some references are not relative to LLN or ULN and are declared like this:
.. code-block:: python
grading_data = {
ldl=[
Formula(
"4.90<=x",
grade=3,
units=MILLIMOLES_PER_LITER,
gender=[MALE, FEMALE],
**adult_age_options,
fasting=True,
),
],
...
}
See edc_reportable.data.grading_data for a complete example.
Note: If the lower and upper values of a grade reference overlap
with another grade reference in the same group, a BoundaryOverlap
exception will be raised when the value is evaluated.
Catch this in your tests.
Important:
Writing out references is prone to error. It is better to declare a
dictionary of normal references and grading references as shown above. Use the Formula class
so that you can use a phrase like 13.5<=x<=17.5 instead of a listing attributes.
Attempting to grade a value without grading data ++++++++++++++++++++++++++++++++++++++++++++++++ If a value is pased to the evaluator and no grading data exists in the reference lists for that test, an exception is raised.
Limiting what is "gradeable" for your project
+++++++++++++++++++++++++++++++++++++++++++++
The default tables have grading data for grades 1-4. The evaluator will grade any value
if there is grading data. You can prevent the evaluator from considering grades by passing
reportable_grades when you register the normal and grading data.
For example, in your reportables.py:
.. code-block:: python
...
reportable_grades = [3, 4]
...
In the above, by explicitly passing a list of grades, the evaluator will only raise an exception for grades 3 and 4. If a value meets the criteria for grade 1 or 2, it will be ignored.
Declaring minor exceptions ++++++++++++++++++++++++++
Minor exceptions can be specified using the parameter reportable_grades_exceptions.
For example, you wish to report grades 2,3,4 for Serum Amylase
but grades 3,4 for everything else. You would register as follows:
.. code-block:: python
...
reportable_grades_exceptions={"amylase": [GRADE2, GRADE3, GRADE4]}
...
Exporting the reference tables ++++++++++++++++++++++++++++++
You can export your declared references to CSV for further inspection using the management command
.. code-block:: python
python manage.py export_reportables
('/Users/erikvw/my_project_normal_data.csv',
'/Users/erikvw/my_project_grading_data.csv')
.. |pypi| image:: https://img.shields.io/pypi/v/edc-reportable.svg :target: https://pypi.python.org/pypi/edc-reportable
.. |actions| image:: https://github.com/clinicedc/edc-reportable/actions/workflows/build.yml/badge.svg :target: https://github.com/clinicedc/edc-reportable/actions/workflows/build.yml
.. |codecov| image:: https://codecov.io/gh/clinicedc/edc-reportable/branch/develop/graph/badge.svg :target: https://codecov.io/gh/clinicedc/edc-reportable
FAQs
Reportable clinic events, reference ranges, grading for clinicedc/edc projects
We found that edc-reportable demonstrated a healthy version release cadence and project activity because the last version was released less than a year ago. It has 2 open source maintainers collaborating on the project.
Did you know?

Socket for GitHub automatically highlights issues in each pull request and monitors the health of all your open source dependencies. Discover the contents of your packages and block harmful activity before you install or update your dependencies.

Security News
The Rust Security Response WG is warning of phishing emails from rustfoundation.dev targeting crates.io users.
Product
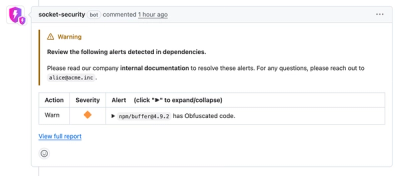
Socket now lets you customize pull request alert headers, helping security teams share clear guidance right in PRs to speed reviews and reduce back-and-forth.

Product
Socket's Rust support is moving to Beta: all users can scan Cargo projects and generate SBOMs, including Cargo.toml-only crates, with Rust-aware supply chain checks.