
Research
/Security News
Critical Vulnerability in NestJS Devtools: Localhost RCE via Sandbox Escape
A flawed sandbox in @nestjs/devtools-integration lets attackers run code on your machine via CSRF, leading to full Remote Code Execution (RCE).
🚀 GO3 is a high-performance Rust library with Python bindings for calculating semantic similarity between Gene Ontology (GO) terms and gene products. Designed to be significantly faster and more memory-efficient than traditional Python libraries like goatools, GO3 provides state-of-the-art similarity measures including Resnik, Lin, Jiang-Conrath, SimRel, GraphIC, Information Coefficient, and Wang.
✅ Ultra-fast ontology loading (50x faster than Goatools)
✅ Parallel computation of semantic similarity (Resnik, Lin, Jiang-Conrath, SimRel, GraphIC, IC Coefficient, Wang)
✅ Both term-to-term and gene-to-gene similarity
✅ Supports batch processing of large datasets
✅ Full compatibility with Gene Association Files (GAF)
✅ Low memory footprint thanks to Rust's performance
pip install go3
✅ Requires Python 3.8+ ✅ Rust is bundled via maturin, no manual compilation needed
import go3
# Load Gene Ontology (GO) terms and annotations
go_terms = go3.load_go_terms()
annotations = go3.load_gaf("goa_human.gaf")
# Build IC Counter
counter = go3.build_term_counter(annotations)
# Compute Resnik similarity between two GO terms
sim = go3.semantic_similarity("GO:0008150", "GO:0009987", 'resnik', counter)
print(f"Resnik similarity: {sim:.4f}")
# Compute similarity between two genes using Lin and Best-Match Average (BMA)
score = go3.compare_genes("TP53", "BRCA1", "BP", "lin", "bma", counter)
print(f"Gene similarity (Lin, BMA): {score:.4f}")
| Measure | Type | Reference |
|---|---|---|
| Resnik | IC-based | Resnik, 1995 |
| Lin | IC-based | Lin, 1998 |
| Jiang-Conrath | IC-based | Jiang & Conrath, 1997 |
| SimRel | IC-based | Schlicker et al., 2006 |
| GraphIC | Hybrid | Li et al., 2010 |
| IC Coefficient | Hybrid | Li et al., 2010 |
| Wang | Topology | Wang et al., 2007 |
| TopoICSim | Hybrid | Ehsani et al., 2016 |
For the theoretical details behind each measure, see the Similarity Measures Documentation.
GO3 natively supports efficient parallel batch computations for both term and gene similarity.
pairs = [("GO:0008150", "GO:0009987"), ("GO:0008150", "GO:0003674")]
scores = go3.batch_similarity([a for a, _ in pairs], [b for _, b in pairs], "resnik", counter)
gene_pairs = [("TP53", "BRCA1"), ("EGFR", "AKT1")]
scores = go3.compare_gene_pairs_batch(gene_pairs, "BP", "resnik", "bma", counter)
Both resnik and lin (and all other similarity methods) are fully supported in batch mode.
This library is built as fast, scalable and memory-efficient as possible. Comparing with Goatools, which is the de facto library for manipulating GO in Python
We compare the time and peak memory consumption of go3 vs goatools while loading the ontology and the annotation (.GAF) file, and building the TermCounter.

We also compare the speed of the libraries calculating the similarities between batches of GO Terms of different sizes.

Finally, we compare the gene similarity calculation times. Goatools does not implement natively the groupwise algorithms to compare genes, so we built it for a fair comparison in top of the GO term semantic similarities of the library.

We welcome contributions!
Steps to contribute:
git checkout -b feature/my-feature
pytest tests/
MIT License © Jose Luis Mellina Andreu, 2025
📄 Full documentation: https://go3.readthedocs.io
🐞 Report issues: https://github.com/Mellandd/go3/issues
FAQs
Semantic similarity computations using Gene Ontology with Rust performance
We found that go3 demonstrated a healthy version release cadence and project activity because the last version was released less than a year ago. It has 1 open source maintainer collaborating on the project.
Did you know?

Socket for GitHub automatically highlights issues in each pull request and monitors the health of all your open source dependencies. Discover the contents of your packages and block harmful activity before you install or update your dependencies.

Research
/Security News
A flawed sandbox in @nestjs/devtools-integration lets attackers run code on your machine via CSRF, leading to full Remote Code Execution (RCE).
Product
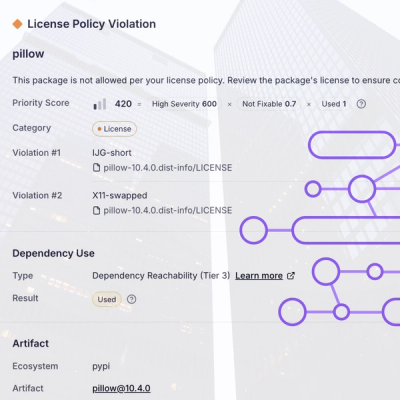
Customize license detection with Socket’s new license overlays: gain control, reduce noise, and handle edge cases with precision.

Product
Socket now supports Rust and Cargo, offering package search for all users and experimental SBOM generation for enterprise projects.