
Research
/Security News
Critical Vulnerability in NestJS Devtools: Localhost RCE via Sandbox Escape
A flawed sandbox in @nestjs/devtools-integration lets attackers run code on your machine via CSRF, leading to full Remote Code Execution (RCE).
To generate a word2vec model, but using multi-word keywords instead of single words.
A simple and fast way to generate a word2vec model, with multi-word keywords instead of single words.
Finding similar keywords for "obesity"
| index | term |
|---|---|
| 0 | overweight |
| 1 | obese |
| 2 | physical inactivity |
| 3 | excess weight |
| 4 | obese adults |
| 5 | high bmi |
| 6 | obese adults |
| 7 | obese people |
| 8 | obesity-related outcomes |
| 9 | obesity among children |
| 10 | poor sleep quality |
| 11 | ssbs |
| 12 | obese populations |
| 13 | cardiometabolic risk |
| 14 | abdominal obesity |
pip install keywords2vec
Lets download some example data
data_filepath = "epistemonikos_data_sample.tsv.gz"
!wget "https://s3.amazonaws.com/episte-labs/epistemonikos_data_sample.tsv.gz" -O "{data_filepath}"
We create the model. If you need the vectors, take a look here
labels, tree = similars_tree(data_filepath)
processing file: epistemonikos_data_sample.tsv.gz
Then we can get the most similars keywords
get_similars(tree, labels, "obesity")
['obesity',
'overweight',
'obese',
'physical inactivity',
'excess weight',
'high bmi',
'obese adults',
'obese people',
'obesity-related outcomes',
'obesity among children',
'poor sleep quality',
'ssbs',
'obese populations',
'cardiometabolic risk',
'abdominal obesity']
get_similars(tree, labels, "heart failure")
['heart failure',
'hf',
'chf',
'chronic heart failure',
'reduced ejection fraction',
'unstable angina',
'peripheral vascular disease',
'peripheral arterial disease',
'angina',
'congestive heart failure',
'left ventricular systolic dysfunction',
'acute coronary syndrome',
'heart failure patients',
'acute myocardial infarction',
'left ventricular dysfunction']
The idea started in the Epistemonikos database www.epistemonikos.org, a database of scientific articles for people making decisions concerning clinical or health-policy questions. In this context the scientific/health language used is complex. You can easily find keywords like:
We tried some approaches to find those keywords, like ngrams, ngrams + tf-idf, identify entities, among others. But we didn't get really good results.
We found that tokenizing using stopwords + non word characters was really useful for "finding" the keywords. An example:
So we basically split the text when we find:
That's it.
But as there were some problem with some keywords that cointain stopwords, like:
So we decided to add another method (nltk with some grammar definition) to cover most of the cases. To use this, you need to add the parameter keywords_w_stopwords=True, this method is approx 20x slower.
Seem to be an old idea (2004):
Mihalcea, Rada, and Paul Tarau. "Textrank: Bringing order into text." Proceedings of the 2004 conference on empirical methods in natural language processing. 2004.
Reading an implementation of textrank, I realize they used stopwords to separate and create the graph. Then I though in using it as tokenizer for word2vec
As pointed by @deliprao in this twitter thread. It's also used by Rake (2010):
Rose, Stuart & Engel, Dave & Cramer, Nick & Cowley, Wendy. (2010). Automatic Keyword Extraction from Individual Documents. 10.1002/9780470689646.ch1.
As noted by @astent in the Twitter thread, this concept is called chinking (chunking by exclusion) https://www.nltk.org/book/ch07.html#Chinking
We worked in an implementation, that could be used in multiple languages. Of course not all languages are sutable for using this approach. We have tried with good results in English, Spanish and Portuguese
You can try it here (takes time to load, lowercase only, doesn't work in mobile yet) MPV :)
These embedding were created using 827,341 title/abstract from @epistemonikos database. With keywords that repeat at least 10 times. The total vocab is 349,080 keywords (really manageable number)
One of the main benefit of this method, is the size of the vocabulary. For example, using keywords that repeat at least 10 times, for the Epistemonikos dataset (827,341 title/abstract), we got the following vocab size:
| ngrams | keywords | comp |
|---|---|---|
| 1 | 127,824 | 36% |
| 1,2 | 1,360,550 | 388% |
| 1-3 | 3,204,099 | 914% |
| 1-4 | 4,461,930 | 1,272% |
| 1-5 | 5,133,619 | 1,464% |
| stopword tokenizer | 350,529 | 100% |
More information regarding the comparison, take a look to the folder analyze.
This project has been created using nbdev
FAQs
To generate a word2vec model, but using multi-word keywords instead of single words.
We found that keywords2vec demonstrated a healthy version release cadence and project activity because the last version was released less than a year ago. It has 1 open source maintainer collaborating on the project.
Did you know?

Socket for GitHub automatically highlights issues in each pull request and monitors the health of all your open source dependencies. Discover the contents of your packages and block harmful activity before you install or update your dependencies.

Research
/Security News
A flawed sandbox in @nestjs/devtools-integration lets attackers run code on your machine via CSRF, leading to full Remote Code Execution (RCE).
Product
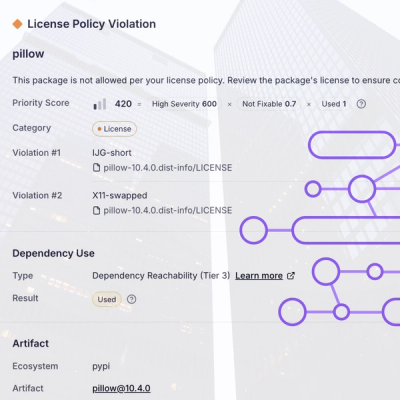
Customize license detection with Socket’s new license overlays: gain control, reduce noise, and handle edge cases with precision.

Product
Socket now supports Rust and Cargo, offering package search for all users and experimental SBOM generation for enterprise projects.