Security News
Crates.io Users Targeted by Phishing Emails
The Rust Security Response WG is warning of phishing emails from rustfoundation.dev targeting crates.io users.
kgenealogic is a tool for genetic genealogy. It clusters relatives into a hypothesized family
tree based on shared and unshared DNA.
It is highly recommended to use a virtual environment for installing kgenealogic and its dependencies (e.g., conda, venv, etc.). Within the virtual environment, just run:
pip install kgenealogic
python3 -m kgenealogic init [-p <project-file>]
python3 -m kgenealogic add [-p <project-file>] ... # files to import
python3 -m kgenealogic cluster [-p <project-file>] [-o <out-file>] <config-file>
python3 -m kgenealogic --help # general help
python3 -m kgenealogic <command> --help # help for <command>, e.g. init/add/build/cluster
Clustering is performed by recursive greedy approximation of a min-cut of a genetic closeness graph.
Specifically, the sums of the (cM) lengths of pairwise matches form the edge-weights of a base graph. We add to these weights the positive and negative lengths of triangulations for which the "source" kit of the triangle is listed as a "seed" in the clustering configuration file. When processing a particular node of the tree, we use triangulations for seeds at that node or on its path to the root (descendants when viewed as a family tree, ancestors when viewed as a tree data structure).
Having formed this graph at a particular node of the tree, we consider seeds listed in the clustering configuration in the maternal or paternal branches of the node. For each connected component of the graph, if there is at least one seed present in the component, we find an approximate minimum cut separating the maternal and paternal seeds using a greedy algorithm. If there are only, e.g., maternal seeds present and no negative weights (from imputed negative triangulations), then the entire component will be classified as maternal.
Kits that are classified as either maternal or paternal at a particular node are considered recursively in the maternal or paternal branches of that mode.
Suppose triangulations and pairwise matches are available for a kit S. And suppose there are kits T1 and T2 that each match pairwise with kit S on a particular segment, but fail to triangulate with S on that segment. Then we have a negative triangulation for source S between T1 and T2, indicating that T1 and T2 likely belong to different branches of the tree. This is represented with a negative weight between T1 and T2 in the graph used for clustering.
We find all negative triangulations for kits S whose triangulations and pairwise matches are available. This is somewhat computationally expensive, because the segments where negative triangulations occur are generally sub-segments of those appearing in the data files.
FAQs
Genetic genealogy tool for tree structure inference
We found that kgenealogic demonstrated a healthy version release cadence and project activity because the last version was released less than a year ago. It has 1 open source maintainer collaborating on the project.
Did you know?

Socket for GitHub automatically highlights issues in each pull request and monitors the health of all your open source dependencies. Discover the contents of your packages and block harmful activity before you install or update your dependencies.

Security News
The Rust Security Response WG is warning of phishing emails from rustfoundation.dev targeting crates.io users.
Product
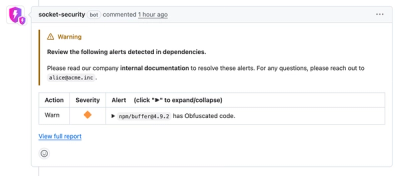
Socket now lets you customize pull request alert headers, helping security teams share clear guidance right in PRs to speed reviews and reduce back-and-forth.

Product
Socket's Rust support is moving to Beta: all users can scan Cargo projects and generate SBOMs, including Cargo.toml-only crates, with Rust-aware supply chain checks.