
Research
wget to Wipeout: Malicious Go Modules Fetch Destructive Payload
Socket's research uncovers three dangerous Go modules that contain obfuscated disk-wiping malware, threatening complete data loss.
Mappy provides a convenient interface to minimap2 <https://github.com/lh3/minimap2>_, a fast and accurate C program to align
genomic and transcribe nucleotide sequences.
Mappy depends on zlib <http://zlib.net>. It can be installed with pip <https://en.wikipedia.org/wiki/Pip_(package_manager)>:
.. code:: shell
pip install --user mappy
or from the minimap2 github repo (Cython <http://cython.org>_ required):
.. code:: shell
git clone https://github.com/lh3/minimap2
cd minimap2
python setup.py install
The following Python script demonstrates the key functionality of mappy:
.. code:: python
import mappy as mp
a = mp.Aligner("test/MT-human.fa") # load or build index
if not a: raise Exception("ERROR: failed to load/build index")
s = a.seq("MT_human", 100, 200) # retrieve a subsequence from the index
print(mp.revcomp(s)) # reverse complement
for name, seq, qual in mp.fastx_read("test/MT-orang.fa"): # read a fasta/q sequence
for hit in a.map(seq): # traverse alignments
print("{}\t{}\t{}\t{}".format(hit.ctg, hit.r_st, hit.r_en, hit.cigar_str))
Mappy implements two classes and two global function.
Class mappy.Aligner
.. code:: python
mappy.Aligner(fn_idx_in=None, preset=None, ...)
This constructor accepts the following arguments:
* **fn_idx_in**: index or sequence file name. Minimap2 automatically tests the
file type. If a sequence file is provided, minimap2 builds an index. The
sequence file can be optionally gzip'd. This option has no effect if **seq**
is set.
* **seq**: a single sequence to index. The sequence name will be set to
:code:`N/A`.
* **preset**: minimap2 preset. Currently, minimap2 supports the following
presets: **sr** for single-end short reads; **map-pb** for PacBio
read-to-reference mapping; **map-ont** for Oxford Nanopore read mapping;
**splice** for long-read spliced alignment; **asm5** for assembly-to-assembly
alignment; **asm10** for full genome alignment of closely related species. Note
that the Python module does not support all-vs-all read overlapping.
* **k**: k-mer length, no larger than 28
* **w**: minimizer window size, no larger than 255
* **min_cnt**: mininum number of minimizers on a chain
* **min_chain_score**: minimum chaing score
* **bw**: chaining and alignment band width (initial chaining and extension)
* **bw_long**: chaining and alignment band width (RMQ-based rechaining and closing gaps)
* **best_n**: max number of alignments to return
* **n_threads**: number of indexing threads; 3 by default
* **extra_flags**: additional flags defined in minimap.h
* **fn_idx_out**: name of file to which the index is written. This parameter
has no effect if **seq** is set.
* **scoring**: scoring system. It is a tuple/list consisting of 4, 6 or 7
positive integers. The first 4 elements specify match scoring, mismatch
penalty, gap open and gap extension penalty. The 5th and 6th elements, if
present, set long-gap open and long-gap extension penalty. The 7th sets a
mismatch penalty involving ambiguous bases.
.. code:: python
mappy.Aligner.map(seq, seq2=None, cs=False, MD=False)
This method aligns :code:`seq` against the index. It is a generator, *yielding*
a series of :code:`mappy.Alignment` objects. If :code:`seq2` is present, mappy
performs paired-end alignment, assuming the two ends are in the FR orientation.
Alignments of the two ends can be distinguished by the :code:`read_num` field
(see Class mappy.Alignment below). Argument :code:`cs` asks mappy to generate
the :code:`cs` tag; :code:`MD` is similar. These two arguments might slightly
degrade performance and are not enabled by default.
.. code:: python
mappy.Aligner.seq(name, start=0, end=0x7fffffff)
This method retrieves a (sub)sequence from the index and returns it as a Python
string. :code:`None` is returned if :code:`name` is not present in the index or
the start/end coordinates are invalid.
.. code:: python
mappy.Aligner.seq_names
This property gives the array of sequence names in the index.
Class mappy.Alignment
This class describes an alignment. An object of this class has the following properties:
ctg: name of the reference sequence the query is mapped to
ctg_len: total length of the reference sequence
r_st and r_en: start and end positions on the reference
q_st and q_en: start and end positions on the query
strand: +1 if on the forward strand; -1 if on the reverse strand
mapq: mapping quality
blen: length of the alignment, including both alignment matches and gaps but excluding ambiguous bases.
mlen: length of the matching bases in the alignment, excluding ambiguous base matches.
NM: number of mismatches, gaps and ambiguous positions in the alignment
trans_strand: transcript strand. +1 if on the forward strand; -1 if on the reverse strand; 0 if unknown
is_primary: if the alignment is primary (typically the best and the first to generate)
read_num: read number that the alignment corresponds to; 1 for the first read and 2 for the second read
cigar_str: CIGAR string
cigar: CIGAR returned as an array of shape :code:(n_cigar,2). The two
numbers give the length and the operator of each CIGAR operation.
MD: the :code:MD tag as in the SAM format. It is an empty string unless
the :code:MD argument is applied when calling :code:mappy.Aligner.map().
cs: the :code:cs tag.
An :code:Alignment object can be converted to a string with :code:str() in
the following format:
::
q_st q_en strand ctg ctg_len r_st r_en mlen blen mapq cg:Z:cigar_str
It is effectively the PAF format without the QueryName and QueryLength columns (the first two columns in PAF).
Miscellaneous Functions
.. code:: python
mappy.fastx_read(fn, read_comment=False)
This generator function opens a FASTA/FASTQ file and *yields* a
:code:`(name,seq,qual)` tuple for each sequence entry. The input file may be
optionally gzip'd. If :code:`read_comment` is True, this generator yields
a :code:`(name,seq,qual,comment)` tuple instead.
.. code:: python
mappy.revcomp(seq)
Return the reverse complement of DNA string :code:`seq`. This function
recognizes IUB code and preserves the letter cases. Uracil :code:`U` is
complemented to :code:`A`.
FAQs
Minimap2 python binding
We found that mappy demonstrated a healthy version release cadence and project activity because the last version was released less than a year ago. It has 1 open source maintainer collaborating on the project.
Did you know?

Socket for GitHub automatically highlights issues in each pull request and monitors the health of all your open source dependencies. Discover the contents of your packages and block harmful activity before you install or update your dependencies.

Research
Socket's research uncovers three dangerous Go modules that contain obfuscated disk-wiping malware, threatening complete data loss.
Research
Socket uncovers malicious packages on PyPI using Gmail's SMTP protocol for command and control (C2) to exfiltrate data and execute commands.
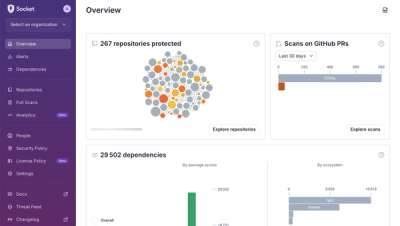
Product
We redesigned Socket's first logged-in page to display rich and insightful visualizations about your repositories protected against supply chain threats.