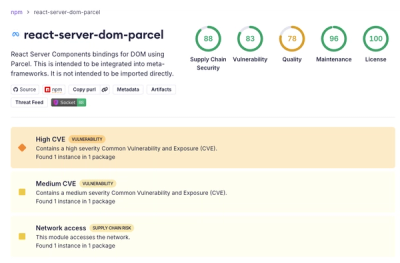
Security News
New React Server Components Vulnerabilities: DoS and Source Code Exposure
New DoS and source code exposure bugs in React Server Components and Next.js: what’s affected and how to update safely.
mavehgvs
Advanced tools
Regular expression-based validation of HGVS-style variant strings for Multiplexed Assays of Variant Effect.
mavehgvs is the Python reference implementation of the MAVE-HGVS variant representation standard, a strict subset of HGVS, used primarily for clinical genomics.
MAVE-HGVS is a strict subset of the HGVS Sequence Variant Nomenclature, version 20.05. HGVS nomenclature is comprehensive and very expressive and consequently includes a lot of syntax that is not needed to represent variants from Multiplexed Assay of Variant Effect (MAVE) data and makes the variant strings more challenging to parse.
While packages exist for parsing HGVS (most notably the biocommons hgvs package, they are intended for use in human genetics and rely on sequence databases and reference sequence (called "target sequence" for MAVE-HGVS), which are not always available for or relevant for multiplexed assays.
MAVE-HGVS is an attempt to define an easy-to-parse subset of the HGVS nomenclature that captures those variants that occur in MAVE datasets, while excluding many variant types that are unlikely to be found. Importantly, the mavehgvs implementation does not rely on external sequence databases or identifiers.
MAVE-HGVS supports DNA, RNA, and protein variants. MAVE-HGVS supports a subset of HGVS variants including:
Many HGVS variants are unsupported including:
For further details, including example variants, see the specification in the package documentation.
Install mavehgvs from pip using:
pip3 install mavehgvs
To set up the package for development purposes, include the optional dependencies and install pre-commit:
pip3 install mavehgvs[dev]
pre-commit install
To report a problem or request a new feature with either the mavehgvs package or the MAVE-HGVS standard, please use the GitHub issue tracker.
FAQs
Regular expression-based validation of HGVS-style variant strings for Multiplexed Assays of Variant Effect.
We found that mavehgvs demonstrated a healthy version release cadence and project activity because the last version was released less than a year ago. It has 2 open source maintainers collaborating on the project.
Did you know?

Socket for GitHub automatically highlights issues in each pull request and monitors the health of all your open source dependencies. Discover the contents of your packages and block harmful activity before you install or update your dependencies.

Security News
New DoS and source code exposure bugs in React Server Components and Next.js: what’s affected and how to update safely.

Security News
Socket CEO Feross Aboukhadijeh joins Software Engineering Daily to discuss modern software supply chain attacks and rising AI-driven security risks.

Security News
GitHub has revoked npm classic tokens for publishing; maintainers must migrate, but OpenJS warns OIDC trusted publishing still has risky gaps for critical projects.