
Research
/Security News
Critical Vulnerability in NestJS Devtools: Localhost RCE via Sandbox Escape
A flawed sandbox in @nestjs/devtools-integration lets attackers run code on your machine via CSRF, leading to full Remote Code Execution (RCE).
Mitosis is an experiment runner. It handles administrative tasks to decrease the mental overhead of collaboration:
The virtuous consequence of these checks and organization is a faster workflow, a more rigorous scientific method, and reduced mental overhead of collaboration.
Hypothesis: the maximum value of a sine wave is equal to its amplitude.
sine_experiment/__init__.py
import numpy as np
import matplotlib.pyplot as plt
name = "sine-exp"
lookup_dict = {"frequency": {"fast": 10, "slow": 1}}
def run(amplitude, frequency):
"""Deterimne if the maximum value of the sine function equals ``amplitude``"""
x = np.arange(0, 10, .05)
y = amplitude * np.sin(frequency * x)
err = np.abs(max(y) - amplitude)
plt.title("What's the maximum value of a sine wave?")
plt.plot(x, y, label="trial data")
plt.plot(x, amplitude * np.ones_like(x), label="expected")
plt.legend()
return {"main": err, "data": y}
pyproject.toml
[tool.mitosis.steps]
my_exp = ["sine_experiment:run", "sine_experiment:lookup_dict"]
Commit these changes to a repository. After installing sine_experiment as a python package, in CLI, run:
mitosis my_exp --param my_exp.frequency=slow --eval-param my_exp.amplitude=4
Mitosis will run sin_experiment.run(), saving
all output as an html file in a subdirectory. It will also
track the parameters and results.
If you later change the variant named "slow" to set frequency=2, mitosis will
raise a RuntimeError, preventing you from running a trial. If you want to run
sine_experiment with a different parameter value, you need to name that variant
something new. Eval parameters, like "amplitude" in the example, behave differently.
Rather than being specified by lookup_dict, they are evaluated directly.
Philosophically, an experiment is any time we run code with an aim to convince someone of something. As code, mitosis takes the approach that an experiment is a callable (or a sequence of callables).
Using mitosis involves registering experiments in pyproject.toml, naming interesting parameters, running experiments on the command line, and browsing results.
mitosis uses the tool.mitosis.steps table of pyproject.toml to learn
what python callables are experiment steps
and where to lookup named parameter values.
It uses a syntax evocative of entry points:
[tool.mitosis.steps]
my_exp = ["sine_experiment:run", "sine_experiment:lookup_dict"]
Experiment steps must be callables with a dictionary return type. The returned dictionary is required to have a key "main". All but the final step in an experiment must also have a key "data" that gets passed to the first argument of the subsequent step. If the key "metrics" is present, it will display prominently in the HTML output
Developer note: Building an experiment step static type at mitosis._typing.ExpRun
The basic invocation lists the steps along with the values of any parameters for each step.
mitosis [OPTION...] step [steps...] [[-p step.lookup_param=key...]
[-e step.eval_param=val...]]...
Some nuance:
--debug can be used to waive a lot of the reproducibility checks mitosis does.
This arg allows you to run experiments in a dirty git repository (or no repository)
and will neither save results in the experimental database, nor increment the trials
counter, nor verify/lock in the definitions of any variants. It will, however,
create the output notebook. It also changes the experiment log level from INFO
to DEBUG.+ to the parameter, e.g. -e +jax_playground.gpu_id=1.-e smoothing.kernel=\"rbf\")-e and -p are short form for --eval-param and --param (lookup param).Trials are saved in trials/ (or whatever is passed after -F). Each trial has a
pseudorandom bytes key, postpended to a metadata folder and an html output filename.
There are two obviously useful things to do after an experiment:
python -m http.server is helpful to browse resultsmitosis.load_trial_data()Beyond this, the metadata mitosis keeps to disk is useful for troubleshooting or reproducing experiments, but no facility yet exists to browse or compare experiments.
Mitosis is primarily intended as a command line program, so mitosis --help has the syntax documentation.
There is only one intentionally public part of the api: mitosis.load_trial_data().
FAQs
Reproduce Machine Learning experiments easily
We found that mitosis demonstrated a healthy version release cadence and project activity because the last version was released less than a year ago. It has 1 open source maintainer collaborating on the project.
Did you know?

Socket for GitHub automatically highlights issues in each pull request and monitors the health of all your open source dependencies. Discover the contents of your packages and block harmful activity before you install or update your dependencies.

Research
/Security News
A flawed sandbox in @nestjs/devtools-integration lets attackers run code on your machine via CSRF, leading to full Remote Code Execution (RCE).
Product
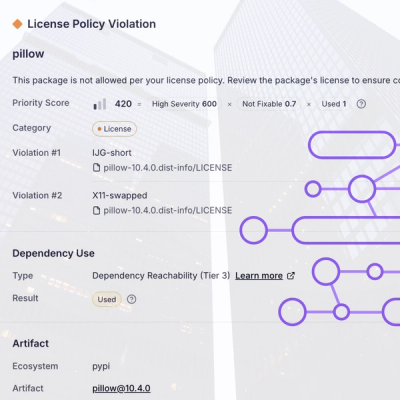
Customize license detection with Socket’s new license overlays: gain control, reduce noise, and handle edge cases with precision.

Product
Socket now supports Rust and Cargo, offering package search for all users and experimental SBOM generation for enterprise projects.