Security News
Crates.io Users Targeted by Phishing Emails
The Rust Security Response WG is warning of phishing emails from rustfoundation.dev targeting crates.io users.
A small Python wrapper for setting up Quantum Espresso input files. More functionality may be added later.
Currently, there is only assumed support for hexagonal close packed (HCP) crystal structures. Support for other structures may be tested but should not be expected.
Note: The
calc.create_bash()function (described later) assumes a Torque/PBS structure and sets up script to run in parallel using MPI. It also assumes an already built Quantum Espresso package and thatpw.xcan be run from the command-line.For tips on how to install Quantum Espresso on an HPC cluster, go to Espresso Installation.

Ninia is a genus of snakes, also known as coffee snakes, that are native to parts of Central and South America. The choice of name is a play on Python and Quantum Espresso.
# Import necessary modules:
from ase.build import molecule, add_adsorbate, hcp0001
from ase.visualize import view
# Set up geometry using ASE:
surface = hcp0001('Ru', size=(4, 2, 4), a=2.7059, c=4.2815)
ad = molecule('NH2')
ad.rotate(180, 'x')
add_adsorbate(surface, ad, 2.0, 'hcp')
view(surface, viewer='x3d') # Specific viewer for use in Jupyter
This will display a view of the geometry we have created. More information about ASE (Atomic Simulation Environment) can be found at their homepage: https://wiki.fysik.dtu.dk/ase/
Then you can start using ninia to convert this geometry into an input file:
from ninia import relax
calc = relax.Relax(prefix='Ru_test', functional='beef')
calc.set_directories(outputdir='/home/ajs0201/workQE/output',
pseudodir='/home/ajs0201/workQE/pseudo')
# Ninia assumes the current script directory as the input directory
# if none is given.
calc.load_geometry(surface)
calc.set_parameters(mixing_beta=0.15)
calc.create_input()
calc.create_job(hours=20)
# This will create both an input (.i) file and bash (.sh) for the geometry above
If you do not specify the prefix and functional during the initialization step, the program will give warnings. Additionally, the pseudopotential directory must be set before the calc.load_geometry() step.
In the calc.set_directories() step, you can set the following directories:
Note: The output directory, post calculation, will contain wave function files (.wfc) and save directories (.save/). This does not include output files (.out), which will be place in the input directory, unless explicitly changed.
In the calc.set_parameters() step, you can set the following parameters [default]:
In the calc.create_job() step, you can set the following parameters [default]:
pbs.sh] (other option slurm.sh)general]FAQs
A small Python wrapper for Quantum Espresso - still in development
We found that ninia demonstrated a healthy version release cadence and project activity because the last version was released less than a year ago. It has 1 open source maintainer collaborating on the project.
Did you know?

Socket for GitHub automatically highlights issues in each pull request and monitors the health of all your open source dependencies. Discover the contents of your packages and block harmful activity before you install or update your dependencies.

Security News
The Rust Security Response WG is warning of phishing emails from rustfoundation.dev targeting crates.io users.
Product
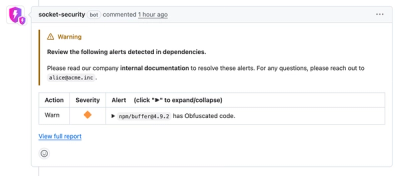
Socket now lets you customize pull request alert headers, helping security teams share clear guidance right in PRs to speed reviews and reduce back-and-forth.

Product
Socket's Rust support is moving to Beta: all users can scan Cargo projects and generate SBOMs, including Cargo.toml-only crates, with Rust-aware supply chain checks.