
Security News
curl Shuts Down Bug Bounty Program After Flood of AI Slop Reports
A surge of AI-generated vulnerability reports has pushed open source maintainers to rethink bug bounties and tighten security disclosure processes.
@gmod/bam
Advanced tools
$ npm install --save @gmod/bam
const { BamFile } = require('@gmod/bam')
// or import {BamFile} from '@gmod/bam'
const t = new BamFile({
bamPath: 'test.bam',
})
// note: it's required to first run getHeader before any getRecordsForRange
var header = await t.getHeader()
// this would get same records as samtools view ctgA:1-50000
var records = await t.getRecordsForRange('ctgA', 0, 50000)
The bamPath argument only works on nodejs. In the browser, you should pass
bamFilehandle with a generic-filehandle2 e.g. RemoteFile
const { RemoteFile } = require('generic-filehandle2')
const bam = new BamFile({
bamFilehandle: new RemoteFile('yourfile.bam'), // or a full http url
baiFilehandle: new RemoteFile('yourfile.bam.bai'), // or a full http url
})
Input are 0-based half-open coordinates (note: not the same as samtools view coordinate inputs!)
Since 1.0.41 we support usage of the htsget protocol
Here is a small code snippet for this
const { HtsgetFile } = require('@gmod/bam')
const ti = new HtsgetFile({
baseUrl: 'http://htsnexus.rnd.dnanex.us/v1/reads',
trackId: 'BroadHiSeqX_b37/NA12878',
})
await ti.getHeader()
const records = await ti.getRecordsForRange(1, 2000000, 2000001)
Our implementation makes some assumptions about how the protocol is implemented, so let us know if it doesn't work for your use case
The BAM class constructor accepts arguments
bamPath/bamUrl/bamFilehandle - a string file path to a local file or a
class object with a read methodcsiPath/csiUrl/csiFilehandle - a CSI index for the BAM file, required
for long chromosomes greater than 2^29 in lengthbaiPath/baiUrl/baiFilehandle - a BAI index for the BAM filerecordClass - a custom class extending BamRecord to use for records (see
Custom BamRecord class section below)Note: filehandles implement the Filehandle interface from https://www.npmjs.com/package/generic-filehandle2.
This module offers the path and url arguments as convenience methods for supplying the LocalFile and RemoteFile
Note: you must run getHeader before running getRecordsForRange
refName - a string for the chrom to fetch fromstart - a 0-based half open start coordinateend - a 0-based half open end coordinateopts.signal - an AbortSignal to indicate stop processingopts.viewAsPairs - re-dispatches requests to find mate pairs. default: falseopts.pairAcrossChr - control the viewAsPairs option behavior to pair across
chromosomes. default: falseopts.maxInsertSize - control the viewAsPairs option behavior to limit
distance within a chromosome to fetch. default: 200kbThis is a async generator function that takes the same signature as
getRecordsForRange but results can be processed using
for await (const chunk of file.streamRecordsForRange(
refName,
start,
end,
opts,
)) {
}
The getRecordsForRange simply wraps this process by concatenating chunks into
an array
This obtains the header from HtsgetFile or BamFile. Retrieves BAM file and
BAI/CSI header if applicable, or API request for refnames from htsget
refName - a string for the chrom to fetch fromstart - a 0-based half open start coordinate (optional)end - a 0-based half open end coordinate (optional)Returns features of the form {start, end, score} containing estimated feature density across 16kb windows in the genome
refName - a string for the chrom to fetch fromReturns number of features on refName, uses special pseudo-bin from the BAI/CSI index (e.g. bin 37450 from bai, returning n_mapped from SAM spec pdf) or -1 if refName not exist in sample
refName - a string for the chrom to checkReturns whether we have this refName in the sample
// Core alignment fields
record.fileOffset // "file offset" based id -- not a true file offset
record.ref_id // numerical sequence id from SAM header
record.start // 0-based start coordinate
record.end // 0-based end coordinate
record.name // QNAME
record.seq // sequence string
record.qual // Uint8Array of quality scores (null if unmapped)
record.CIGAR // CIGAR string e.g. "50M2I48M"
record.flags // SAM flags integer
record.mq // mapping quality (undefined if 255)
record.strand // 1 or -1
record.template_length // TLEN
// Auxiliary data
record.tags // object with all aux tags e.g. {MD: "100", NM: 0}
record.NUMERIC_MD // MD tag as Uint8Array (for fast mismatch rendering)
record.NUMERIC_CIGAR // Uint32Array of packed CIGAR operations
record.NUMERIC_SEQ // Uint8Array of packed sequence (4-bit encoded)
// Mate info
record.next_refid // mate reference id
record.next_pos // mate position
// Flag methods
record.isPaired()
record.isProperlyPaired()
record.isSegmentUnmapped()
record.isMateUnmapped()
record.isReverseComplemented()
record.isMateReverseComplemented()
record.isRead1()
record.isRead2()
record.isSecondary()
record.isFailedQc()
record.isDuplicate()
record.isSupplementary()
// Utility
record.seqAt(idx) // get single base at position
record.toJSON() // serialize record
You can provide your own BamRecord class to add custom properties or methods:
import { BamFile, BamRecord } from '@gmod/bam'
class CustomBamRecord extends BamRecord {
get customProperty() {
return `custom-${this.name}`
}
getDoubleStart() {
return this.start * 2
}
}
const bam = new BamFile<CustomBamRecord>({
bamPath: 'test.bam',
recordClass: CustomBamRecord,
})
await bam.getHeader()
const records = await bam.getRecordsForRange('ctgA', 0, 50000)
// records are typed as CustomBamRecord[]
console.log(records[0].customProperty)
console.log(records[0].getDoubleStart())
MIT © Colin Diesh
FAQs
Parser for BAM and BAM index (bai) files
The npm package @gmod/bam receives a total of 946 weekly downloads. As such, @gmod/bam popularity was classified as not popular.
We found that @gmod/bam demonstrated a healthy version release cadence and project activity because the last version was released less than a year ago. It has 6 open source maintainers collaborating on the project.
Did you know?

Socket for GitHub automatically highlights issues in each pull request and monitors the health of all your open source dependencies. Discover the contents of your packages and block harmful activity before you install or update your dependencies.

Security News
A surge of AI-generated vulnerability reports has pushed open source maintainers to rethink bug bounties and tighten security disclosure processes.
Product
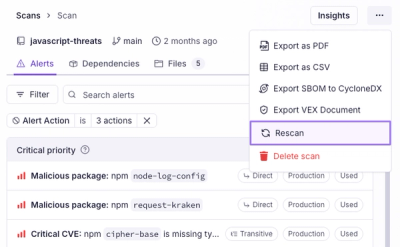
Scan results now load faster and remain consistent over time, with stable URLs and on-demand rescans for fresh security data.
Product
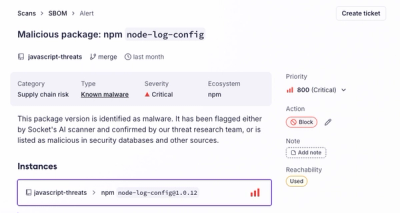
Socket's new Alert Details page is designed to surface more context, with a clearer layout, reachability dependency chains, and structured review.