Product
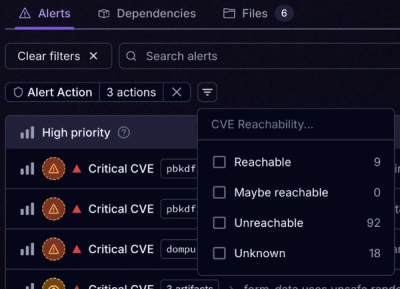
Introducing Tier 1 Reachability: Precision CVE Triage for Enterprise Teams
Socket’s new Tier 1 Reachability filters out up to 80% of irrelevant CVEs, so security teams can focus on the vulnerabilities that matter.
.. image:: https://badge.fury.io/py/biothings-client.svg :target: https://pypi.python.org/pypi/biothings-client
.. image:: https://img.shields.io/pypi/pyversions/biothings-client.svg :target: https://pypi.python.org/pypi/biothings-client
.. image:: https://img.shields.io/pypi/format/biothings-client.svg :target: https://pypi.python.org/pypi/biothings-client
.. image:: https://github.com/biothings/biothings_client.py/actions/workflows/build-package.yml/badge.svg :target: https://github.com/biothings/biothings_client.py/actions/workflows/build-package.yml
.. image:: https://github.com/biothings/biothings_client.py/actions/workflows/run-tests.yml/badge.svg :target: https://github.com/biothings/biothings_client.py/actions/workflows/run-tests.yml
.. image:: https://api.codacy.com/project/badge/Grade/2b716becb6e948a6b576f278e64dd81f :alt: Codacy Badge :target: https://www.codacy.com/gh/biothings/biothings_client.py/dashboard?utm_source=github.com&utm_medium=referral&utm_content=biothings/biothings_client.py&utm_campaign=Badge_Grade
.. image:: https://app.codacy.com/project/badge/Coverage/2b716becb6e948a6b576f278e64dd81f :alt: Codacy Coverage Badge :target: https://app.codacy.com/gh/biothings/biothings_client.py/dashboard?utm_source=gh&utm_medium=referral&utm_content=&utm_campaign=Badge_coverage
.. image:: https://img.shields.io/pypi/dm/biothings_client.svg :alt: PyPI Downloads :target: https://pypistats.org/packages/biothings_client
.. image:: https://readthedocs.org/projects/biothings-clientpy/badge/?version=latest :alt: Documentation Status :target: https://biothings-clientpy.readthedocs.io/en/latest/?badge=latest
biothings_client is an easy-to-use Python wrapper to access any Biothings.api_ -based backend service. Currently, the following clients are available:
* gene - The client for MyGene.Info_, which provides access to gene objects.
* variant - The client MyVariant.Info_, which provides access to genetic variant objects.
* chem - The client for MyChem.Info_, which provides access to chemical/drug objects.
* disease - The client for MyDisease.Info_, which provides access to disease objects.
* geneset - The client for MyGeneset.Info_, which provides access to geneset/pathway objects.
* taxon - The client for t.biothings.io_, which provides access to taxon objects.
.. _t.biothings.io: https://t.biothings.io .. _Biothings.api: https://biothings.io .. _MyGene.Info: https://mygene.info .. _MyVariant.Info: https://myvariant.info .. _MyChem.Info: https://mychem.info .. _MyDisease.Info: https://mydisease.info .. _MyGeneset.Info: https://mygeneset.info
python >=3.7
* `pandas <http://pandas.pydata.org>`_ (install using "pip install pandas") is required for returning a list of variant objects as `DataFrame <http://pandas.pydata.org/pandas-docs/stable/dsintro.html#dataframe>`_.
* Option 1. Install from pypi repository using pip:
.. code-block:: bash
pip install biothings_client
* Option 2. Install the latest commit from the github repository using pip:
.. code-block:: bash
pip install -e git+https://github.com/biothings/biothings_client.py#egg=biothings_client
`CHANGES.txt <https://raw.githubusercontent.com/biothings/biothings_client.py/master/CHANGES.txt>`_
See the `quick start tutorial <https://biothings-clientpy.readthedocs.io/en/latest/doc/Quick-Start.html>`_ at the `biothings_client doc page <https://biothings-clientpy.readthedocs.io/en/latest/index.html>`_.
https://biothings-clientpy.readthedocs.io
Synchronous client.
.. code-block:: python
from biothings_client import get_client
# get a client for variant objects
mv = get_client("variant")
mv.getvariant("chr7:g.140453134T>C")
# output below is collapsed
{"_id": "chr7:g.140453134T>C",
"_version": 1,
"chrom": "7",
"cadd": {...},
"clinvar": {...},
"cosmic": {...},
"dbnsfp": {...},
"dbsnp": {...},
"docm": {...},
"hg19": {'end': 140453134, 'start': 140453134},
"mutdb": {...},
"snpeff": {...},
"vcf": {
"alt": "C",
"position": "140453134",
"ref": "T"
}}
# get a client for gene objects
mg = get_client("gene")
mg.getgene(1017, 'name,symbol,refseq')
{'_id': '1017',
'_score': 21.03413,
'name': 'cyclin dependent kinase 2',
'refseq': {'genomic': ['NC_000012.12', 'NC_018923.2', 'NG_034014.1'],
'protein': ['NP_001277159.1',
'NP_001789.2',
'NP_439892.2',
'XP_011536034.1'],
'rna': ['NM_001290230.1', 'NM_001798.4', 'NM_052827.3', 'XM_011537732.1'],
'translation': [{'protein': 'NP_001789.2', 'rna': 'NM_001798.4'},
{'protein': 'NP_439892.2', 'rna': 'NM_052827.3'},
{'protein': 'NP_001277159.1', 'rna': 'NM_001290230.1'},
{'protein': 'XP_011536034.1', 'rna': 'XM_011537732.1'}]},
'symbol': 'CDK2'}
# get a client for chems/drugs
md = get_client("chem")
md.getchem("ATBDZSAENDYQDW-UHFFFAOYSA-N", fields="pubchem")
{'_id': 'ATBDZSAENDYQDW-UHFFFAOYSA-N',
'_version': 1,
'pubchem': {'chiral_atom_count': 0,
'chiral_bond_count': 0,
'cid': 'CID4080429',
'complexity': 250,
'covalently-bonded_unit_count': 1,
'defined_atom_stereocenter_count': 0,
'defined_bond_stereocenter_count': 0,
'exact_mass': 184.019415,
'formal_charge': 0,
'heavy_atom_count': 12,
'hydrogen_bond_acceptor_count': 3,
'hydrogen_bond_donor_count': 1,
'inchi': 'InChI=1S/C8H8O3S/c1-2-7-4-3-5-8(6-7)12(9,10)11/h2-6H,1H2,(H,9,10,11)',
'inchi_key': 'ATBDZSAENDYQDW-UHFFFAOYSA-N',
'isotope_atom_count': 0,
'iupac': {'traditional': '3-vinylbesylic acid'},
'molecular_formula': 'C8H8O3S',
'molecular_weight': 184.21232,
'monoisotopic_weight': 184.019415,
'rotatable_bond_count': 2,
'smiles': {'isomeric': 'C=CC1=CC(=CC=C1)S(=O)(=O)O'},
'tautomers_count': 1,
'topological_polar_surface_area': 62.8,
'undefined_atom_stereocenter_count': 0,
'undefined_bond_stereocenter_count': 0,
'xlogp': 1.4}}
# get a client for taxa
mt = get_client("taxon")
mt.gettaxon(9606)
{'_id': '9606',
'_version': 1,
'authority': ['homo sapiens linnaeus, 1758'],
'common_name': 'man',
'genbank_common_name': 'human',
'has_gene': True,
'lineage': [9606,
9605,
207598,
9604,
314295,
9526,
314293,
376913,
9443,
314146,
1437010,
9347,
32525,
40674,
32524,
32523,
1338369,
8287,
117571,
117570,
7776,
7742,
89593,
7711,
33511,
33213,
6072,
33208,
33154,
2759,
131567,
1],
'other_names': ['humans'],
'parent_taxid': 9605,
'rank': 'species',
'scientific_name': 'homo sapiens',
'taxid': 9606,
'uniprot_name': 'homo sapiens'}
Asynchronous client.
.. code-block:: python
from biothings_client import get_async_client
# get a client for variant objects
mv = get_async_client("variant")
await mv.getvariant("chr7:g.140453134T>C")
# get a client for gene objects
mg = get_async_client("gene")
await mg.getgene(1017, 'name,symbol,refseq')
# get a client for chems/drugs
md = get_async_client("chem")
await md.getchem("ATBDZSAENDYQDW-UHFFFAOYSA-N", fields="pubchem")
# get a client for taxa
mt = get_async_client("taxon")
await mt.gettaxon(9606)
Drop us any feedback @biothingsapi <https://twitter.com/biothingsapi>_
FAQs
Python Client for BioThings API services.
We found that biothings-client demonstrated a healthy version release cadence and project activity because the last version was released less than a year ago. It has 2 open source maintainers collaborating on the project.
Did you know?

Socket for GitHub automatically highlights issues in each pull request and monitors the health of all your open source dependencies. Discover the contents of your packages and block harmful activity before you install or update your dependencies.

Product
Socket’s new Tier 1 Reachability filters out up to 80% of irrelevant CVEs, so security teams can focus on the vulnerabilities that matter.
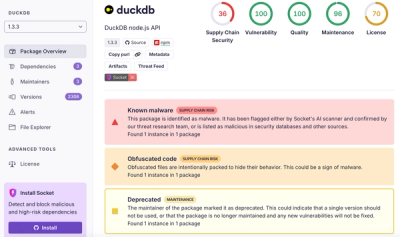
Research
/Security News
Ongoing npm supply chain attack spreads to DuckDB: multiple packages compromised with the same wallet-drainer malware.

Security News
The MCP Steering Committee has launched the official MCP Registry in preview, a central hub for discovering and publishing MCP servers.