Product
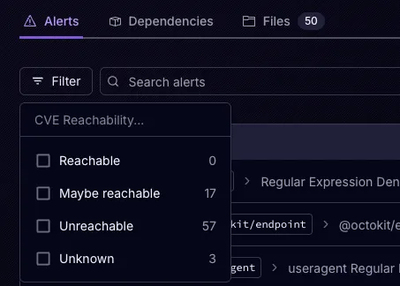
Announcing Precomputed Reachability Analysis in Socket
Socket’s precomputed reachability slashes false positives by flagging up to 80% of vulnerabilities as irrelevant, with no setup and instant results.
capanno-utils is a companion tool to manage metadata and workflow language files for bioinformatics tools in the capanno and similar repositories. Usage examples and additional documentation can be found in that repository. It is recommended to start with the Getting Started documentation.
schema-salad-tool --codegen python https://github.com/common-workflow-language/common-workflow-language/raw/master/v1.0/CommonWorkflowLanguage.yml > capanno_utils/classes/cwl/common_workflow_language.py
or
curl https://raw.githubusercontent.com/common-workflow-language/cwl-utils/main/cwl_utils/parser/cwl_v1_2.py > capanno_utils/classes/cwl/common_workflow_language.py
perl -p -i -e 'print "from capanno_utils.classes.cwl.common_workflow_language_mixins import CommandLineToolMixin, \\
CommandInputParameterMixin, SchemaDefRequirementMixin, CommandLineBindingMixin, \\
CommandOutputParameterMixin, WorkflowMixin, InputParameterMixin, WorkflowOutputParameterMixin, \\
WorkflowStepMixin, WorkflowStepInputMixin\n" if $. == 1' capanno_utils/classes/cwl/common_workflow_language.py
perl -p -i -e 's/SchemaDefRequirement\(/SchemaDefRequirement(SchemaDefRequirementMixin,/; \
s/CommandLineBinding\(/CommandLineBinding(CommandLineBindingMixin,/; \
s/CommandInputParameter\(/CommandInputParameter(CommandInputParameterMixin,/; \
s/CommandOutputParameter\(/CommandOutputParameter(CommandOutputParameterMixin,/; \
s/CommandLineTool\(/CommandLineTool(CommandLineToolMixin,/; \
s/Workflow\(/Workflow(WorkflowMixin,/; \
s/class InputParameter\(/class InputParameter(InputParameterMixin,/; \
s/WorkflowOutputParameter\(/WorkflowOutputParameter(WorkflowOutputParameterMixin,/; \
s/class WorkflowStep\(/class WorkflowStep(WorkflowStepMixin,/; \
s/class WorkflowStepInput\(/class WorkflowStepInput(WorkflowStepInputMixin,/;' capanno_utils/classes/cwl/common_workflow_language.py
Usage:
capanno-add tool kallisto 0.45.x --biotoolsID kallisto #This will initialize a directory for kallisto
tools/
└── kallisto
└── 0.45.x
├── common
│ └── common-metadata.yaml
└── kallisto
├── instances
└── kallisto-metadata.yaml
If you don't provide a biotoolsID, capanno-utils will just make a blank template.
capanno-add tool kallisto 0.45.x
tools/
└── kallisto
└── 0.45.x
└── common
└── common-metadata.yaml
#remote
capanno-add subtool kallisto 0.45.x index -u --init-cwl https://github.com/common-workflow-library/bio-cwl-tools/raw/release/Kallisto/Kallisto-Index.cwl
#local
capanno-add subtool kallisto 0.45.x index -u --init-cwl bio-cwl-tools-submodule/Kallisto/Kallisto-Index.cwl
tools/
└── kallisto
└── 0.45.x
├── common
│ └── common-metadata.yaml
├── kallisto
│ ├── instances
│ └── kallisto-metadata.yaml
└── kallisto_index
├── instances
├── kallisto-index-metadata.yaml
└── kallisto-index.cwl
If --init_cwl is not provided, no cwl file is initialized and capanno-utils will make a blank template.
Can also initialize all subtool directories when first initialize the tool directory but haven't made a way to specify cwl urls to initialize each one doing it that way yet.
capanno-add subtool kallisto 0.45.x index -u
cwl-tools/
└── kallisto
└── 0.45.x
├── common
│ └── common-metadata.yaml
└── kallisto_index
├── instances
└── kallisto-index-metadata.yaml
FAQs
Tool for managing bioinformatics content repositories.
We found that capanno-utils demonstrated a healthy version release cadence and project activity because the last version was released less than a year ago. It has 2 open source maintainers collaborating on the project.
Did you know?

Socket for GitHub automatically highlights issues in each pull request and monitors the health of all your open source dependencies. Discover the contents of your packages and block harmful activity before you install or update your dependencies.

Product
Socket’s precomputed reachability slashes false positives by flagging up to 80% of vulnerabilities as irrelevant, with no setup and instant results.

Product
Socket is launching experimental protection for Chrome extensions, scanning for malware and risky permissions to prevent silent supply chain attacks.

Product
Add secure dependency scanning to Claude Desktop with Socket MCP, a one-click extension that keeps your coding conversations safe from malicious packages.