Product
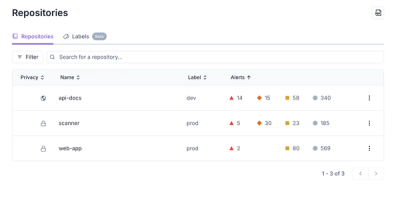
Redesigned Repositories Page: A Faster Way to Prioritize Security Risk
Our redesigned Repositories page adds alert severity, filtering, and tabs for faster triage and clearer insights across all your projects.
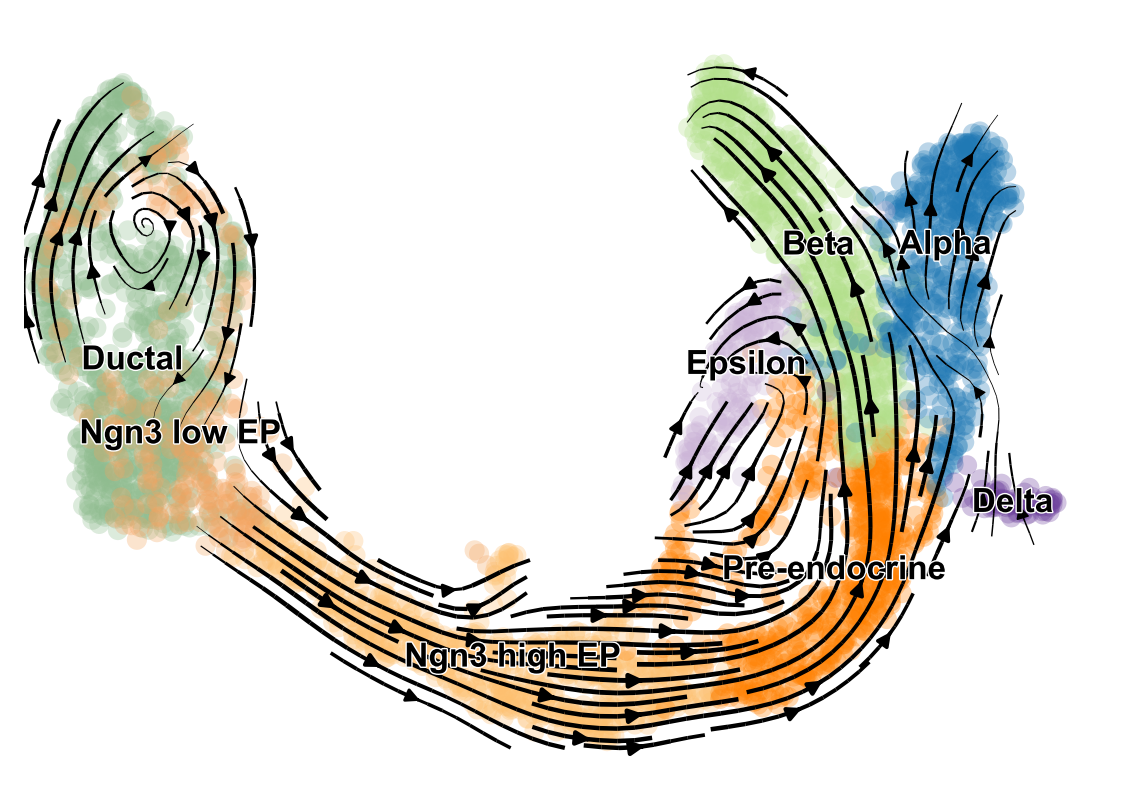
scVelo is a scalable toolkit for RNA velocity analysis in single cells; RNA velocity enables the recovery of directed dynamic information by leveraging splicing kinetics 1. scVelo collects different methods for inferring RNA velocity using an expectation-maximization framework 2, deep generative modeling 3, or metabolically labeled transcripts4.
If you include or rely on scVelo when publishing research, please adhere to the following citation guide:
If you use the EM (dynamical) or steady-state model, cite
@article{Bergen2020,
title = {Generalizing RNA velocity to transient cell states through dynamical modeling},
volume = {38},
ISSN = {1546-1696},
url = {http://dx.doi.org/10.1038/s41587-020-0591-3},
DOI = {10.1038/s41587-020-0591-3},
number = {12},
journal = {Nature Biotechnology},
publisher = {Springer Science and Business Media LLC},
author = {Bergen, Volker and Lange, Marius and Peidli, Stefan and Wolf, F. Alexander and Theis, Fabian J.},
year = {2020},
month = aug,
pages = {1408–1414}
}
If you use the implemented method for estimating RNA velocity from metabolic labeling information, cite
@article{Weiler2024,
author = {Weiler, Philipp and Lange, Marius and Klein, Michal and Pe'er, Dana and Theis, Fabian},
publisher = {Springer Science and Business Media LLC},
url = {http://dx.doi.org/10.1038/s41592-024-02303-9},
doi = {10.1038/s41592-024-02303-9},
issn = {1548-7105},
journal = {Nature Methods},
month = jun,
number = {7},
pages = {1196--1205},
title = {CellRank 2: unified fate mapping in multiview single-cell data},
volume = {21},
year = {2024},
}
Found a bug or would like to see a feature implemented? Feel free to submit an issue. Have a question or would like to start a new discussion? Head over to GitHub discussions. Your help to improve scVelo is highly appreciated. For further information visit scvelo.org.
FAQs
RNA velocity generalized through dynamical modeling
We found that scvelo demonstrated a healthy version release cadence and project activity because the last version was released less than a year ago. It has 2 open source maintainers collaborating on the project.
Did you know?

Socket for GitHub automatically highlights issues in each pull request and monitors the health of all your open source dependencies. Discover the contents of your packages and block harmful activity before you install or update your dependencies.

Product
Our redesigned Repositories page adds alert severity, filtering, and tabs for faster triage and clearer insights across all your projects.

Security News
Slopsquatting is a new supply chain threat where AI-assisted code generators recommend hallucinated packages that attackers register and weaponize.

Security News
Multiple deserialization flaws in PyTorch Lightning could allow remote code execution when loading untrusted model files, affecting versions up to 2.4.0.