Product
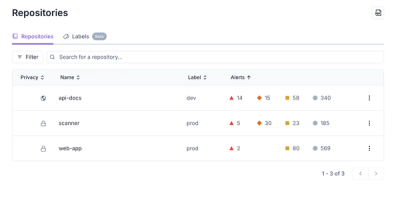
Redesigned Repositories Page: A Faster Way to Prioritize Security Risk
Our redesigned Repositories page adds alert severity, filtering, and tabs for faster triage and clearer insights across all your projects.
synbio is a library for designing and assembling DNA. Users can design plasmids or libraries and export multi-step build protocols. Input SeqRecords. Output assembly SeqRecords, protocols, plate maps, and robotic picklists.
Documentation is available at https://lattice-automation.github.io/synbio
pip install synbio
synbio only expects the user to define their Design and Protocol. Several protocols are pre-defined.
All are in synbio.designs:
Combinatorial - list of SeqRecords to combinatorially anneal into all valid assembliesCombinatorialBins - list of bins of SeqRecords for combinatorial assembly of records between binsPlasmid - single list of SeqRecords to combine into a plasmidPlasmidLibrary - list of list of SeqRecords to combine into plasmidsAll are in synbio.protocols:
Gibson - Gibson assembly based on NEB's E5510GoldenGate - Golden Gate assembly based on NEB's E1601In the example below, the user specifies a combinatorial library design. All SeqRecords are tested for circularization with other SeqRecords. New and valid plasmids are assembled.
Behind the scenes, synbio is filtering all combinations of SeqRecords from the design that will circularize into valid plasmids (via circuits in a graph). After running the protocol, users can export plate maps (to_csv()), composite plasmids (to_fasta(), to_genbank()), and assembly instructions (to_txt(), to_picklists()).
"""Example of a Combinatorial Golden Gate assembly with human and robot output protocols."""
import os
from Bio.SeqIO import parse
from synbio.designs import Combinatorial
from synbio.protocols import GoldenGate
def read_all_records():
gg_dir = os.path.join(".", "data", "goldengate")
records = []
for file in os.listdir(gg_dir):
if file.endswith(".gb"):
records.extend(parse(os.path.join(gg_dir, file), "genbank"))
return records
# create a combinatorial library design from all valid combinations
design = Combinatorial(read_all_records())
# create a protocol using Golden Gate as the sole composite step and run
protocol = GoldenGate(
name="Combinatorial Golden Gate",
design=design,
include=["KanR"], # only keep circularized plasmids with a KanR SeqFeature
min_count=5, # only keep circularized plasmids from >=5 SeqRecords
)
protocol.to_fasta("plasmids.fasta") # export multi-FASTA
protocol.to_csv("plates.csv") # export plate layouts
protocol.to_txt("protocol.txt") # export human protocol
protocol.to_picklists("picklist", platform="hamilton") # export a hamilton picklist
plasmids.fasta:
>J23100_AB+B0032m_BC+C0012m_CD+B0015_DE+DVK_AE
GGAGTTGACGGCTAGCTCAGTCCTAGGTACAGTGCTAGCTACTAGAGTCACACAGGAAAG
TACTAAATGATGGTGAATGTGAAACCAGTAACGTTATACGATGTCGCAGAGTATGCCGGT
...
plates.csv:
Setup Wells with volumes (uL) shown:
Plate:1,1,2,3,4,5,6,7,8,9,10,11,12
A,B0015_DE(4),C0080_CD(18),R0010_AB(54),water(36)
B,B0015_DE(160),DVK_AE(160),cre_CD(18),water(156)
...
protocol.txt:
Combinatorial GoldenGate:
1. Setup PCR plate with (volumes) shown:
1.1. Dilute plasmid DNA to 75 ng/µL in 'water'
1.2. Create 'assembly-mix' from 1:1 T4 Ligase Buffer (10X) and NEB Golden Gate Assembly Mix
...
picklist.gwl:
A;Plate:2;;;15;;2.0;;;
D;Plate:3;;;80;;2.0;;;
W;;;;;;;;;
...
In addition to DNA assembly, synbio exposes a plasmid annotation function in synbio.features. An example below shows a SeqRecord being augmented additional SeqFeatures from a curated database of common plasmid features.
from Bio.SeqIO import parse
from synbio.features import annotate
record = next(parse("plasmid.fa", "fasta"))
record_with_features = annotate(record, identity=0.96)
This is a non-exhaustive list. Contact me for a comparison of these libraries/platforms and synbio.
FAQs
Synbio design and build library
We found that synbio demonstrated a healthy version release cadence and project activity because the last version was released less than a year ago. It has 2 open source maintainers collaborating on the project.
Did you know?

Socket for GitHub automatically highlights issues in each pull request and monitors the health of all your open source dependencies. Discover the contents of your packages and block harmful activity before you install or update your dependencies.

Product
Our redesigned Repositories page adds alert severity, filtering, and tabs for faster triage and clearer insights across all your projects.

Security News
Slopsquatting is a new supply chain threat where AI-assisted code generators recommend hallucinated packages that attackers register and weaponize.

Security News
Multiple deserialization flaws in PyTorch Lightning could allow remote code execution when loading untrusted model files, affecting versions up to 2.4.0.