
Security News
VulnCon 2025: NVD Scraps Industry Consortium Plan, Raising Questions About Reform
At VulnCon 2025, NIST scrapped its NVD consortium plans, admitted it can't keep up with CVEs, and outlined automation efforts amid a mounting backlog.
Tatapov is a Python library making accessible and easy to explore the DNA
overhang misannealing data from
Potapov et al. (2018, ACS Synth. Biol. <https://pubs.acs.org/doi/10.1021/acssynbio.8b00333>)
and Pryor et al. (2020, PLoS ONE <https://doi.org/10.1371/journal.pone.0238592>):
Comprehensive Profiling of Four Base Overhang Ligation Fidelity by T4 DNA Ligase and Application to DNA Assembly. Vladimir Potapov, Jennifer L. Ong, Rebecca B. Kucera, Bradley W. Langhorst, Katharina Bilotti, John M. Pryor, Eric J. Cantor, Barry Canton, Thomas F. Knight, Thomas C. Evans Jr., Gregory Lohman. ACS Synth. Biol. (2018) 7, 11, 2665–2674.
Enabling one-pot Golden Gate assemblies of unprecedented complexity using data-optimized assembly design. John M. Pryor, Vladimir Potapov, Rebecca B. Kucera, Katharina Bilotti, Eric J. Cantor, Gregory J. S. Lohman. PLoS ONE (2020) 15(9): e0238592.
The Supplementary Material of these papers provide tables of inter-overhang annealing data in various conditions (01h or 18h incubations at 25C or 37C). Tatapov provides these tables (it will download them automatically upon first use) as pandas dataframes, so that they are easy to manipulate.
It also provides simple methods to build and plot subsets of the data (plotting requires Matplotlib installed).
Plotting
.. code:: python
import tatapov
data = tatapov.annealing_data["25C"]["01h"] # a pandas dataframe overhangs = ["ACGA", "AAAT", "AGAG"] subset = tatapov.data_subset(data, overhangs, add_reverse=True)
ax, _ = tatapov.plot_data(subset, figwidth=5, plot_color="Blues") ax.figure.tight_layout() ax.figure.savefig("example.png")
.. image:: https://raw.githubusercontent.com/Edinburgh-Genome-Foundry/tatapov/master/images/tatapov_example.png
In the plot above, if you see anything else than the square pairs around the
diagonal, it means there is cross-talking between your overhangs (so risk of misannealing).
If one of these diagonal square pairs appears lighter than the others, it means that
the corresponding overhang has weak self-annealing (risk of having no assembly).
A color square in the diagonal means that the overhang can anneal with itself (palindromic).
The Matplotlib colormap is specified with the plot_color parameter.
The following datasets are available (see the publications for more details):
.. code:: python
tatapov.annealing_data[temperature][time]
tatapov.annealing_data['37C'][enzyme]
Identifying weak self-annealing overhangs
.. code:: python
import tatapov
annealing_data = tatapov.annealing_data['37C']['01h']
# Compute a dictionary {overhang: self-annealing score in 0-1}
relative_self_annealing = tatapov.relative_self_annealings(annealing_data)
weak_self_annealing_overhangs = [
overhang
for overhang, self_annealing in relative_self_annealing.items()
if self_annealing < 0.4
]
Identifying overhang pairs with significant cross-talking
.. code:: python
import tatapov
annealing_data = tatapov.annealing_data['37C']['01h']
# Compute a dictionary {overhang_pair: cross-talking score in 0-1}
cross_annealings = tatapov.cross_annealings(annealing_data)
high_cross_annealing_pairs = [
overhang_pair
for overhang_pair, cross_annealing in cross_annealings.items()
if cross_annealing > 0.08
]
You can install Tatapov through PIP:
.. code::
pip install tatapov
Tatapov is an open-source software originally written at the Edinburgh Genome
Foundry by Zulko <https://github.com/Zulko>_ and
released on Github <https://github.com/Edinburgh-Genome-Foundry/tatapov>_
under the MIT licence (Copyright 2018 Edinburgh Genome Foundry, University of Edinburgh).
.. image:: https://raw.githubusercontent.com/Edinburgh-Genome-Foundry/Edinburgh-Genome-Foundry.github.io/master/static/imgs/logos/egf-codon-horizontal.png :target: https://edinburgh-genome-foundry.github.io/
Tatapov is part of the EGF Codons <https://edinburgh-genome-foundry.github.io/>_
synthetic biology software suite for DNA design, manufacturing and validation.
FAQs
Tools for DNA overhang misannealing data.
We found that tatapov demonstrated a healthy version release cadence and project activity because the last version was released less than a year ago. It has 1 open source maintainer collaborating on the project.
Did you know?

Socket for GitHub automatically highlights issues in each pull request and monitors the health of all your open source dependencies. Discover the contents of your packages and block harmful activity before you install or update your dependencies.

Security News
At VulnCon 2025, NIST scrapped its NVD consortium plans, admitted it can't keep up with CVEs, and outlined automation efforts amid a mounting backlog.
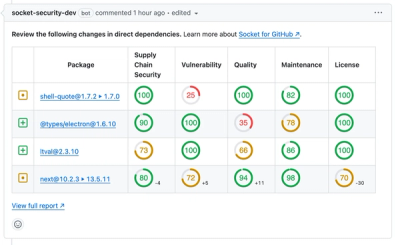
Product
We redesigned our GitHub PR comments to deliver clear, actionable security insights without adding noise to your workflow.
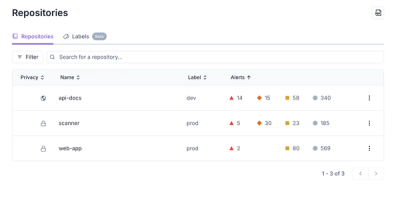
Product
Our redesigned Repositories page adds alert severity, filtering, and tabs for faster triage and clearer insights across all your projects.