
Security News
OWASP 2025 Top 10 Adds Software Supply Chain Failures, Ranked Top Community Concern
OWASP’s 2025 Top 10 introduces Software Supply Chain Failures as a new category, reflecting rising concern over dependency and build system risks.
Vgrid
A unified framework for working with DGGS

Vgrid DGGS is a python package for working with popular geodesic DGGS and graticule-based DGGS.
Vgrid supports a wide range of popular geodesic DGGS, including H3, S2, A5, rHEALPix, Open-EAGGR ISEA4T, ISEA3H, DGGRID, DGGAL, EASE-DGGS, QTM, along with graticule-based DGGS such as OLC, Geohash, MGRS, GEOREF, TileCode, Quadkey, Maidenhead, and GARS.
Vgrid supports popular input and output GIS formats, including CSV, GeoJSON, Pandas/GeoPandas, Shapefile, GeoPackage, and GeoParquet.
The following Vgrid user guide is intended for use in a Command Line Interface (CLI) environment. Full Vgrid DGGS documentation is available at vgrid document.
To work with Vgrid DGGS directly in GeoPandas and Pandas, please use vgridpandas. Full Vgridpandas DGGS documentation is available at vgridpandas document
To work with Vgrid DGGS in QGIS, install the Vgrid Plugin.
To visualize DGGS in Maplibre GL JS, try the vgrid-maplibre library.
For an interactive demo, visit the Vgrid Homepage.
pip install vgrid --upgrade
Convert lat, long in WGS84 CRS to DGGS cellID (H3, S2, A5, rHEALPix, OpenEaggr ISEA4T and ISEA3H, EASE-DGGS, QTM, OLC/ OpenLocationCode/ Google Plus Code, Geohash, GEOREF, MGRS, Tilecode, Quadkey, Maidenhead, GARS).
> latlon2h3 10.775276 106.706797 13 # latlon2h3 <lat> <lon> <resolution> [0..15]
> latlon2s2 10.775276 106.706797 21 # latlon2s2 <lat> <lon> <resolution> [0..30]
> latlon2a5 10.775276 106.706797 18 # latlon2a5 <lat> <lon> <resolution> [0..29]
> latlon2rhealpix 10.775276 106.706797 14 # latlon2rhealpix <lat> <lon> <resolution> [1..15]
> latlon2isea4t 10.775276 106.706797 21 # latlon2isea4t <lat> <lon> <resolution> [0..39]
> latlon2isea3h 10.775276 106.706797 27 # latlon2isea3h <lat> <lon> <resolution> [0..40]
> latlon2ease 10.775276 106.706797 6 # latlon2ease <lat> <lon> <resolution> [0..6]
> latlon2dggrid 10.775276 106.706797 FULLER4D 10 # Linux only: latlon2dggrid <lat> <lon> <dggs_type> <resolution>
> latlon2qtm 10.775276 106.706797 11 # latlon2qtm <lat> <lon> <resolution> [1..24]
> latlon2olc 10.775276 106.706797 11 # latlon2olc <lat> <lon> <resolution> [2,4,6,8,10,11..15]
> latlon2geohash 10.775276 106.706797 9 # latlon2geohash <lat> <lon> <resolution>[1..10]
> latlon2georef 10.775276 106.706797 4 # latlon2georef <lat> <lon> <resolution> [0..10]
> latlon2mgrs 10.775276 106.706797 4 # latlon2mgrs <lat> <lon> <resolution> [0..5]
> latlon2tilecode 10.775276 106.706797 23 # latlon2tilecode <lat> <lon> <resolution> [0..29]
> latlon2quadkey 10.775276 106.706797 23 # latlon2quadkey <lat> <lon> <resolution> [0..29]
> latlon2maidenhead 10.775276 106.706797 4 # latlon2maidenhead <lat> <lon> <resolution> [1..4]
> latlon2gars 10.775276 106.706797 1 # latlon2gars <lat> <lon> <resolution> [1..4] (30,15,5,1 minutes)
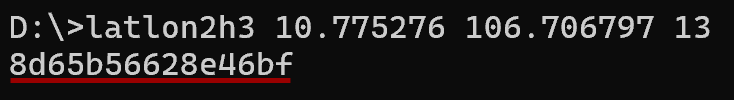
Convert DGGS cell IDs to GeoJSON.
> h32geojson 8d65b56628e46bf
> s22geojson 31752f45cc94
> a52geojson 8e65b56628e0d07
> rhealpix2geojson R31260335553825
> isea4t2geojson 13102313331320133331133
> isea3h2geojson 1327916769,-55086
> ease2geojson L6.165767.02.02.22.45.63.05
> dggrid2geojson 8478420 FULLER4D 10 # Linux only: dggrid2geojson <DGGRID code in SEQNUM format> <DGGS Type> <resolution>
# <DGGS Type> chosen from [SUPERFUND,PLANETRISK,ISEA3H,ISEA4H,ISEA4T,ISEA4D,ISEA43H,ISEA7H,IGEO7,FULLER3H,FULLER4H,FULLER4T,FULLER4D,FULLER43H,FULLER7H]
> qtm2geojson 42012323131
> olc2geojson 7P28QPG4+4P7
> geohash2geojson w3gvk1td8
> mgrs2geojson 34TGK56063228
> gzd # Create Grid Zone Designators - used by MGRS
> tilecode2geojson z23x6680749y3941729
> quadkey2geojson 13223011131020212310000
> maidenhead2geojson OK30is46
> gars2geojson 574JK1918
{"type": "FeatureCollection", "features": [{"type": "Feature", "geometry": {"type": "Polygon", "coordinates": [[[106.70789209957029, 10.77601109480422], [106.70745212203926, 10.777918172217145], [106.7055792173179, 10.778494886227104], [106.70414629547702, 10.777164500398262], [106.70458630070586, 10.775257408234628], [106.70645920007833, 10.774680716650128], [106.70789209957029, 10.77601109480422]]]}, "properties": {"h3": "8965b56628fffff", "resolution": 9, "center_lat": 10.7765878, "center_lon": 106.7060192, "avg_edge_len": 215.295, "cell_area": 120421.396}}]}
Convert Vector layers (Point/ Multipoint, Linestring/ Multilinestring, polygon/ Multipolygon) in GeoJSON to DGGS.
> geojson2h3 -r 11 -geojson polygon.geojson # geojson2h3 -r <resolution>[0..15] -geojson <GeoJSON file> -compact [optional]
> geojson2s2 -r 18 -geojson polygon.geojson -compact # geojson2s2 -r <resolution>[0..30] -geojson <GeoJSON file> -compact [optional]
> geojson2a5 -r 18 -geojson polygon.geojson -compact # geojson2a5 -r <resolution>[0..29] -geojson <GeoJSON file> -compact [optional]
> geojson2rhealpix -r 11 -geojson polygon.geojson # geojson2rhealpix -r <resolution>[1..15] -geojson <GeoJSON file> -compact [optional]
> geojson2isea4t -r 17 -geojson polygon.geojson # geojson2isea4t -r <resolution>[0..25] -geojson <GeoJSON file> -compact [optional]
> geojson2isea3h -r 18 -geojson polygon.geojson # geojson2isea3h -r <resolution>[1..32] -geojson <GeoJSON file> -compact [optional]
> geojson2ease -r 4 -geojson polygon.geojson # geojson2ease -r <resolution>[0..6] -geojson <GeoJSON file> -compact [optional]
> geojson2dggrid -t ISEA4H -r 17 -geojson polyline.geojson # Linux only: geojson2dggrid -t <DGGS Type> -r <resolution> -a <address_type, default is SEQNUM> -geojson <GeoJSON path>
# <Address Type> chosen from [Q2DI,SEQNUM,INTERLEAVE,PLANE,Q2DD,PROJTRI,VERTEX2DD,AIGEN,Z3,Z3_STRING,Z7,Z7_STRING,ZORDER,ZORDER_STRING]
> geojson2qtm -r 18 -geojson polygon.geojson # geojson2qtm -r <resolution>[1..24] -geojson <GeoJSON file> -compact [optional]
> geojson2olc -r 10 -geojson polygon.geojson # geojson2olc -r <resolution>[2,4,6,8,10,11..15] -geojson <GeoJSON file> -compact [optional]
> geojson2geohash -r 7 -geojson polygon.geojson # geojson2geohash -r <resolution>[1..10] -geojson <GeoJSON file> -compact [optional]
> geojson2mgrs -r 3 -geojson polygon.geojson # geojson2mgrs -r <resolution>[0..5] -geojson <GeoJSON file>
> geojson2tilecode -r 18 -geojson polygon.geojson # geojson2tilecode -r <resolution>[0..29] -geojson <GeoJSON file> -compact [optional]
> geojson2quadkey -r 18 -geojson polygon.geojson # geojson2quadkey -r <resolution>[0..29] -geojson <GeoJSON file> -compact [optional]
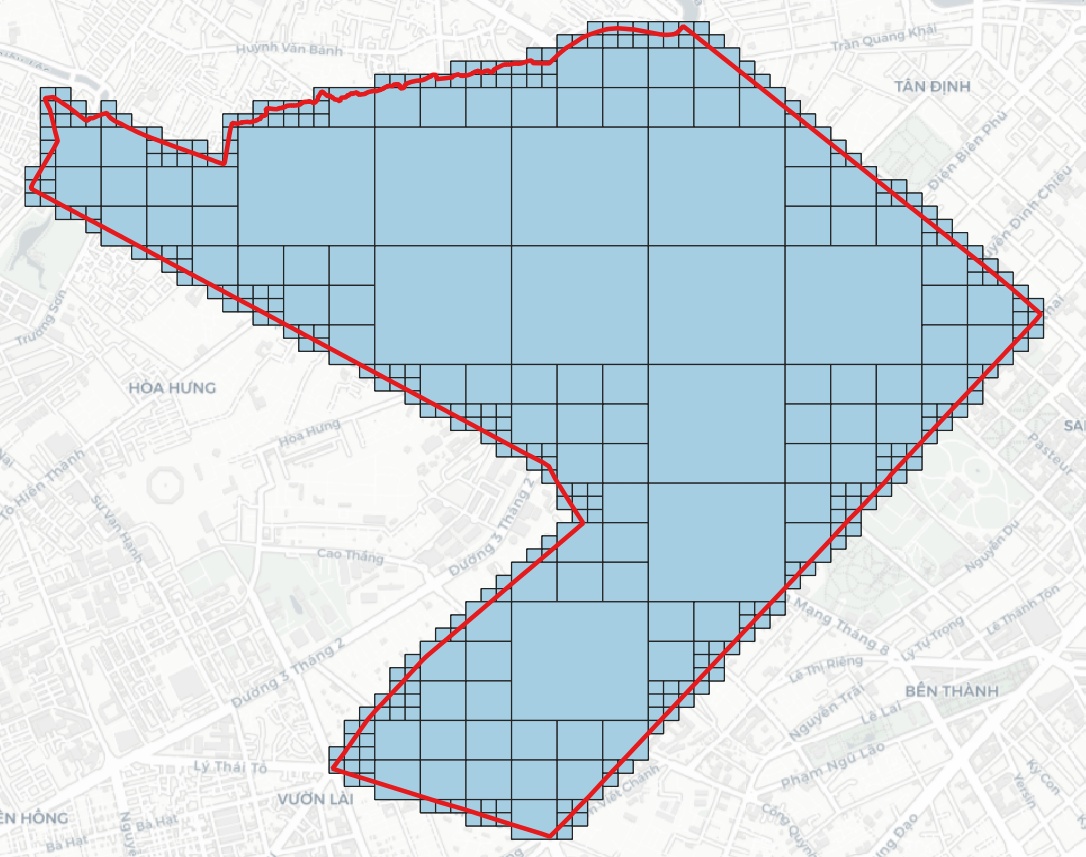
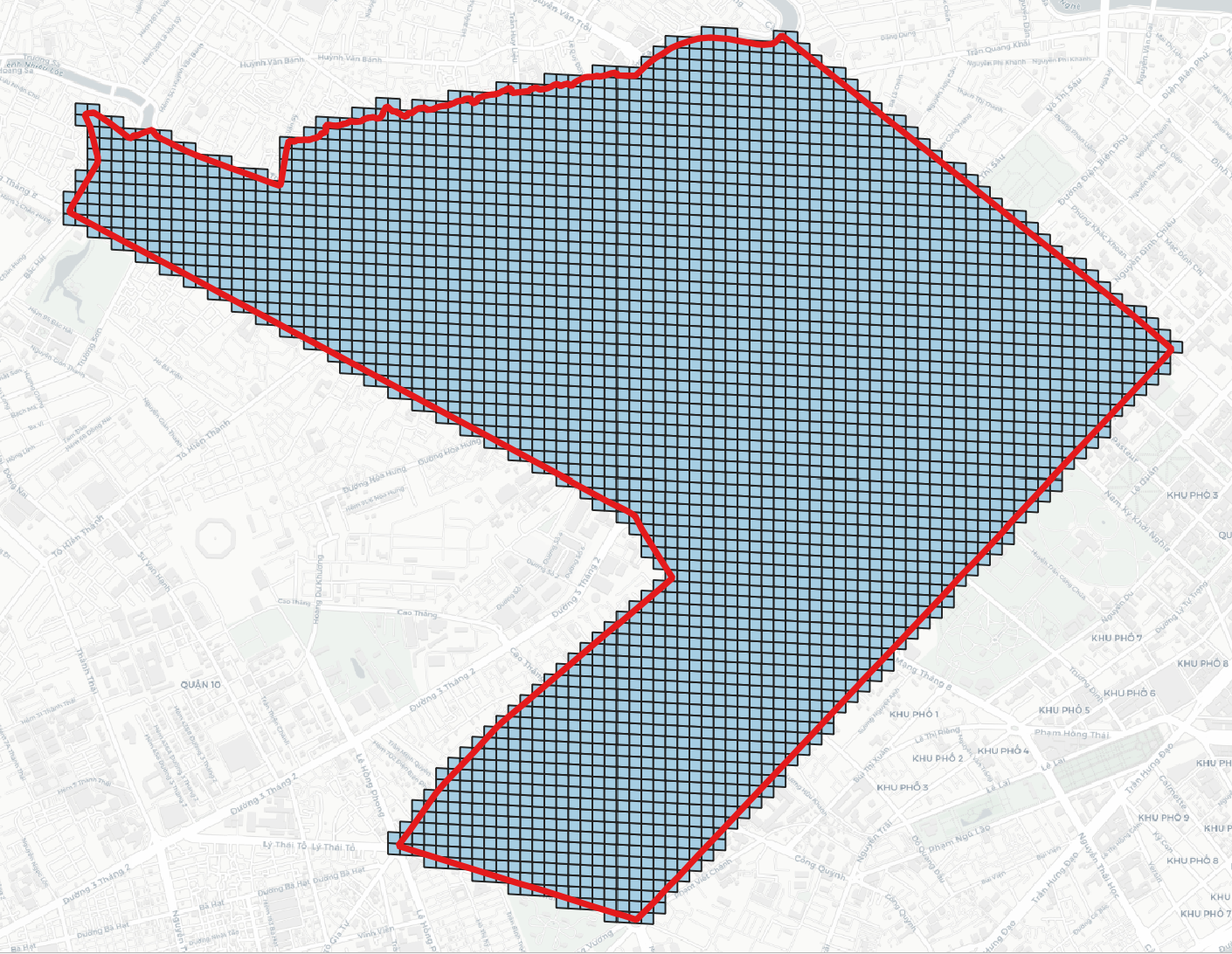
> h3compact -geojson h3.geojson -cellid h3 # h3compact -geojson <H3 in GeoJSON> -cellid [optional, 'h3' by default]
> s2compact -geojson s2.geojson -cellid s2 # s2compact -geojson <S2 in GeoJSON> -cellid [optional, 's2' by default]
> a5compact -geojson a5.geojson -cellid a5 # a5compact -geojson <A5 in GeoJSON> -cellid [optional, 'a5' by default]
> rhealpixcompact -geojson rhealpix.geojson -cellid rhealpix # rhealpixcompact -geojson <rHEALPix in GeoJSON> -cellid [optional, 'rhealpix' by default]
> isea4tcompact -geojson isea4t.geojson -cellid isea4t # isea4tcompact -geojson <ISEA4T in GeoJSON> -cellid [optional, 'isea4t' by default]
> isea3hcompact -geojson isea3h.geojson -cellid isea3h # isea3hcompact -geojson <ISEA3H in GeoJSON> -cellid [optional, 'isea3h' by default]
> easecompact -geojson ease.geojson -cellid ease # easecompact -geojson <EASE in GeoJSON> -cellid [optional, 'ease' by default]
> qtmcompact -geojson qtm.geojson -cellid qtm # qtmcompact -geojson <QTM in GeoJSON> -cellid [optional, 'qtm' by default]
> olccompact -geojson olc.geojson -cellid olc # olccompact -geojson <OLC in GeoJSON> -cellid [optional, 'olc' by default]
> geohashcompact -geojson geohash.geojson -cellid geohash # geohashcompact -geojson <Geohash in GeoJSON> -cellid [optional, 'geohash' by default]
> tilecodecompact -geojson tilecode.geojson -cellid tilecode # tilecodecompact -geojson <Tilecode in GeoJSON> -cellid [optional, 'tilecode' by default]
> quadkeycompact -geojson quadkey.geojson -cellid quadkey # quadkeycompact -geojson <quadkey in GeoJSON> -cellid [optional, 'quadkey' by default]
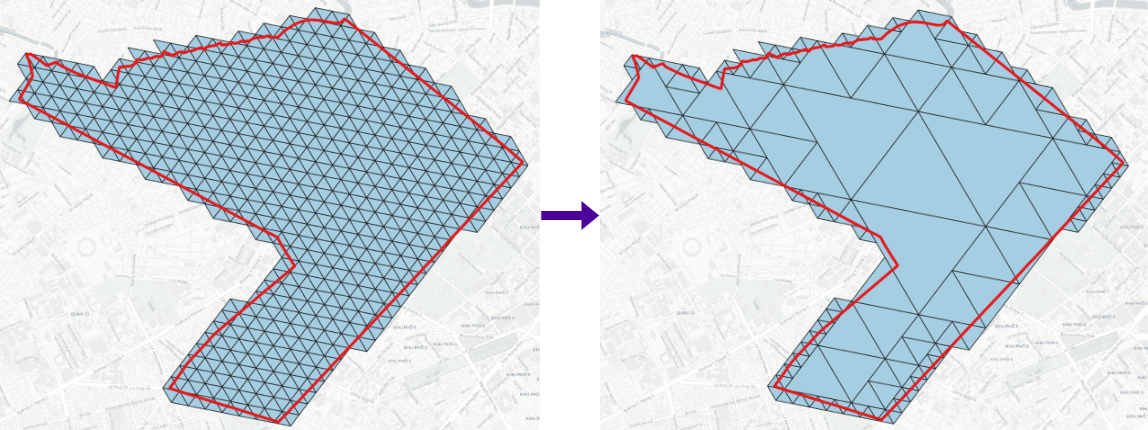
> h3expand -geojson h3_11.geojson -r 12 -cellid h3 # h3expand -geojson <H3 in GeoJSON> -r <higher resolution>[0..15] -cellid [optional, 'h3' by default]
> s2expand -geojson s2_18.geojson -r 19 -cellid s2 # s2expand -geojson <S2 in GeoJSON> -r <higher resolution>[0..30] -cellid [optional, 's2' by default]
> a5expand -geojson a5_18.geojson -r 19 -cellid a5 # a5expand -geojson <A5 in GeoJSON> -r <higher resolution>[0..29] -cellid [optional, 'a5' by default]
> rhealpixexpand -geojson rhealpix_10.geojson -r 11 -cellid rhealpix # rhealpixexpand -geojson <rHEALPix in GeoJSON> -r <higher resolution>[0..15] -cellid [optional, 'rhealpix' by default]
> isea4texpand -geojson isea4t_18.geojson -r 19 -cellid isea4t # isea4texpand -geojson <ISEA4T in GeoJSON> -r <higher resolution>[0..25] -cellid [optional, 'isea4t' by default]
> isea3hexpand -geojson isea3h_18.geojson -r 19 -cellid isea3h # isea3hexpand -geojson <ISEA3H in GeoJSON> -r <higher resolution>[0..25] -cellid [optional, 'isea3h' by default]
> easeexpand -geojson ease_4.geojson -r 5 -cellid ease # easeexpand -geojson <EASE in GeoJSON> -r <higher resolution>[0..6] -cellid [optional, 'ease' by default]
> qtmexpand -geojson qtm_18.geojson -r 19 -cellid qtm # qtmexpand -geojson <QTM in GeoJSON> -r <higher resolution>[1..24] -cellid [optional, 'qtm' by default]
> olcexpand -geojson olc_8.geojson -r 10 -cellid olc # olcexpand -geojson <OLC in GeoJSON> -r <higher resolution>[2,4,6,8,10..15] -cellid [optional, 'olc' by default]
> geohashexpand -geojson geohash_7.geojson -r 8 -cellid geohash # geohashexpand -geojson <Geohash in GeoJSON> -r <higher resolution>[1..10] -cellid [optional, 'geohash' by default]
> tilecodeexpand -geojson tilecode_18.geojson -r 19 -cellid tilecode # tilecodeexpand -geojson <Tilecode in GeoJSON> -r <higher resolution>[0..29] -cellid [optional, 'tilecode' by default]
> quadkeyexpand -geojson quadkey_18.geojson -r 19 -cellid quadkey # quadkeyexpand -geojson <Quadkey in GeoJSON> -r <higher resolution>[0..29] -cellid [optional, 'quadkey' by default]
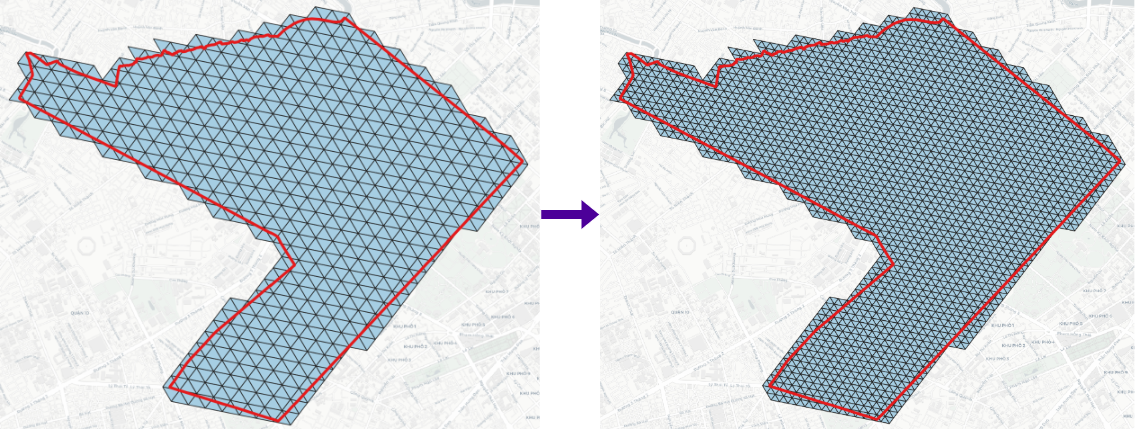
Binning point layer to DGGS
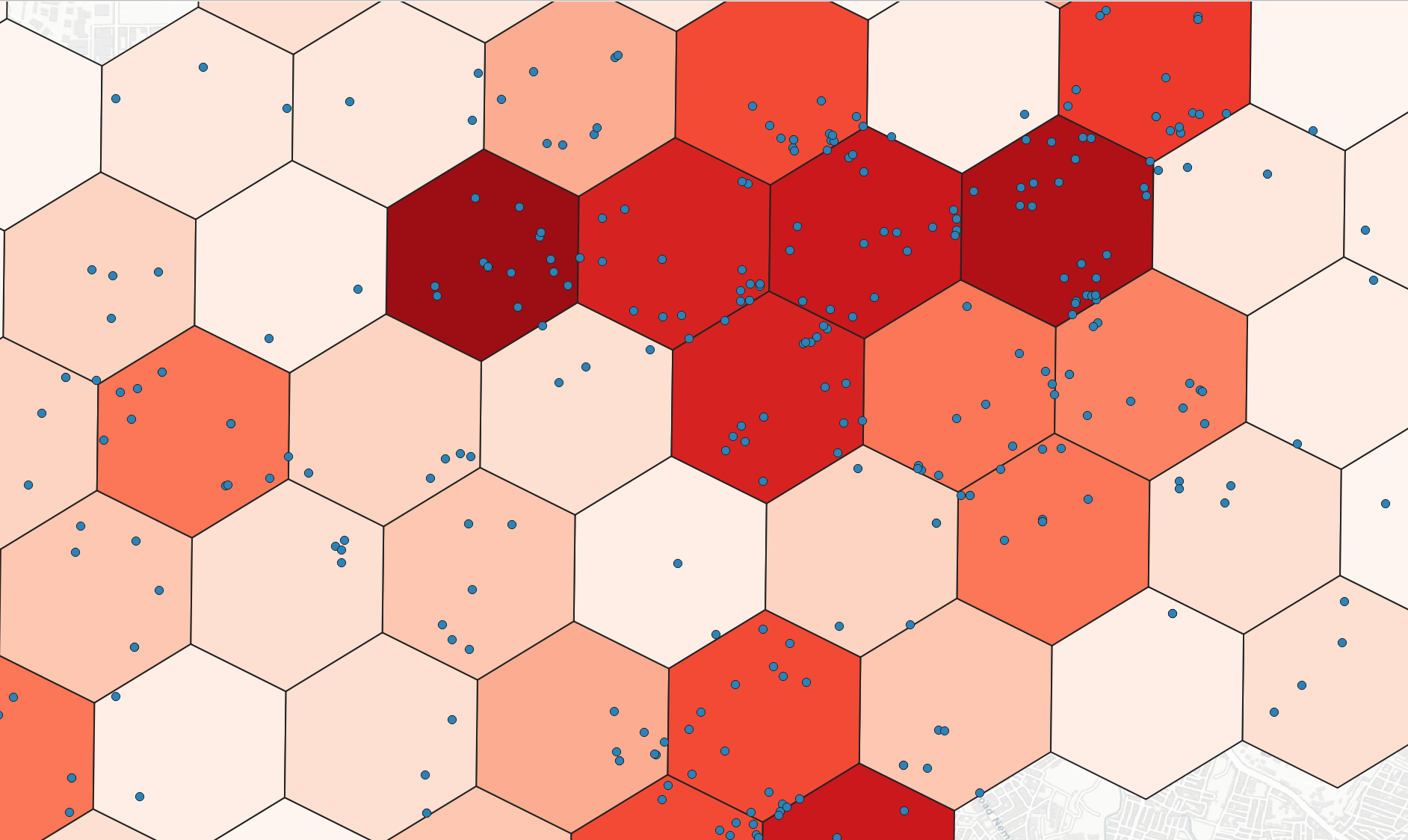
> h3bin -point point.geojson -r 8 -stats count -field numeric_field -category group # h3bin -point <point GeoJSON file> -r <resolution[0..15]> -stats [count, min, max, sum, mean, median, std, var, range, minority, majority, variety] -field [Optional, numeric field to compute statistics] -category [optional, category field for grouping]
> s2bin -point point.geojson -r 13 -stats count -field numeric_field -category group # s2bin -point <point GeoJSON file> -r <resolution[0..30]> -stats [count, min, max, sum, mean, median, std, var, range, minority, majority, variety] -field [Optional, numeric field to compute statistics] -category [optional, category field for grouping]
> a5bin -point point.geojson -r 18 -stats count -field numeric_field -category group # a5bin -point <point GeoJSON file> -r <resolution[0..29]> -stats [count, min, max, sum, mean, median, std, var, range, minority, majority, variety] -field [Optional, numeric field to compute statistics] -category [optional, category field for grouping]
> rhealpixbin -point point.geojson -r 8 -stats count -field numeric_field -category group # rhealpixbin -point <point GeoJSON file> -r <resolution[0..15]> -stats [count, min, max, sum, mean, median, std, var, range, minority, majority, variety] -field [Optional, numeric field to compute statistics] -category [optional, category field for grouping]
> isea4tbin -point point.geojson -r 13 -stats count -field numeric_field -category group # isea4tbin -point <point GeoJSON file> -r <resolution[0..25]> -stats [count, min, max, sum, mean, median, std, var, range, minority, majority, variety] -field [Optional, numeric field to compute statistics] -category [optional, category field for grouping]
> qtmbin -point point.geojson -r 13 -stats count -field numeric_field -category group # qtmbin -point <point GeoJSON file> -r <resolution[1..24]> -stats [count, min, max, sum, mean, median, std, var, range, minority, majority, variety] -field [Optional, numeric field to compute statistics] -category [optional, category field for grouping]
> olcbin -point point.geojson -r 9 -stats count -field numeric_field -category group # olcbin -point <point GeoJSON file> -r <resolution[2,4,6,8,10..15]> -stats [count, min, max, sum, mean, median, std, var, range, minority, majority, variety] -field [Optional, numeric field to compute statistics] -category [optional, category field for grouping]
> geohashbin -point point.geojson -r 6 -stats count -field numeric_field -category group # geohashbin -point <point GeoJSON file> -r <resolution[1..10]> -stats [count, min, max, sum, mean, median, std, var, range, minority, majority, variety] -field [Optional, numeric field to compute statistics] -category [optional, category field for grouping]
> tilecodebin -point point.geojson -r 15 -stats count -field numeric_field -category group # tilecodebin -point <point GeoJSON file> -r <resolutin[0..25]> -stats [count, min, max, sum, mean, median, std, var, range, minority, majority, variety] -field [Optional, numeric field to compute statistics] -category [optional, category field for grouping]
> quadkeybin -point point.geojson -r 13 -stats count -field numeric_field -category group # quadkeybin -point <point GeoJSON file> -r <resolutin[0..25]> -stats [count, min, max, sum, mean, median, std, var, range, minority, majority, variety] -field [Optional, numeric field to compute statistics] -category [optional, category field for grouping]
Convert raster layers in geographic CRS to output DGGS with closest matching resolution.
> raster2h3 -raster raster.tif # raster2h3 -raster <raster in geographic CRS> -r <resolution>[0..15] [optional, defaults to the H3 resolution nearest to the raster's cell size]
> raster2s2 -raster raster.tif # raster2s2 -raster <raster in geographic CRS> -r <resolution>[0..24] [optional, defaults to the S2 resolution nearest to the raster's cell size]
> raster2a5 -raster raster.tif # raster2a5 -raster <raster in geographic CRS> -r <resolution>[0..29] [optional, defaults to the A5 resolution nearest to the raster's cell size]
> raster2rhealpix -raster raster.tif # raster2rhealpix -raster <raster in geographic CRS> -r <resolution>[0..15] [optional, defaults to the rHEALPix resolution nearest to the raster's cell size]
> raster2isea4t -raster raster.tif # raster2isea4t -raster <raster in geographic CRS> -r <resolution>[0..23] [optional, defaults to the ISEA4T resolution nearest to the raster's cell size]
> raster2qtm -raster raster.tif # raster2qtm -raster <raster in geographic CRS> -r <resolution>[1..24] [optional, defaults to the QTM resolution nearest to the raster's cell size]
> raster2olc -raster raster.tif # raster2olc -raster <raster in geographic CRS> -r <resolution>[10..12] [optional, defaults to the OLC resolution nearest to the raster's cell size]
> raster2geohash -raster raster.tif # raster2geohash -raster <raster in geographic CRS> -r <resolution>[1..10] [optional, defaults to the Geohash resolution nearest to the raster's cell size]
> raster2tilecode -raster raster.tif # raster2tilecode -raster <raster in geographic CRS> -r <resolution>[0..26] [optional, defaults to the Tilecode resolution nearest to the raster's cell size]
> raster2quadkey -raster raster.tif # raster2quadkey -raster <raster in geographic CRS> -r <resolution>[0..26] [optional, defaults to the Quadkey resolution nearest to the raster's cell size]

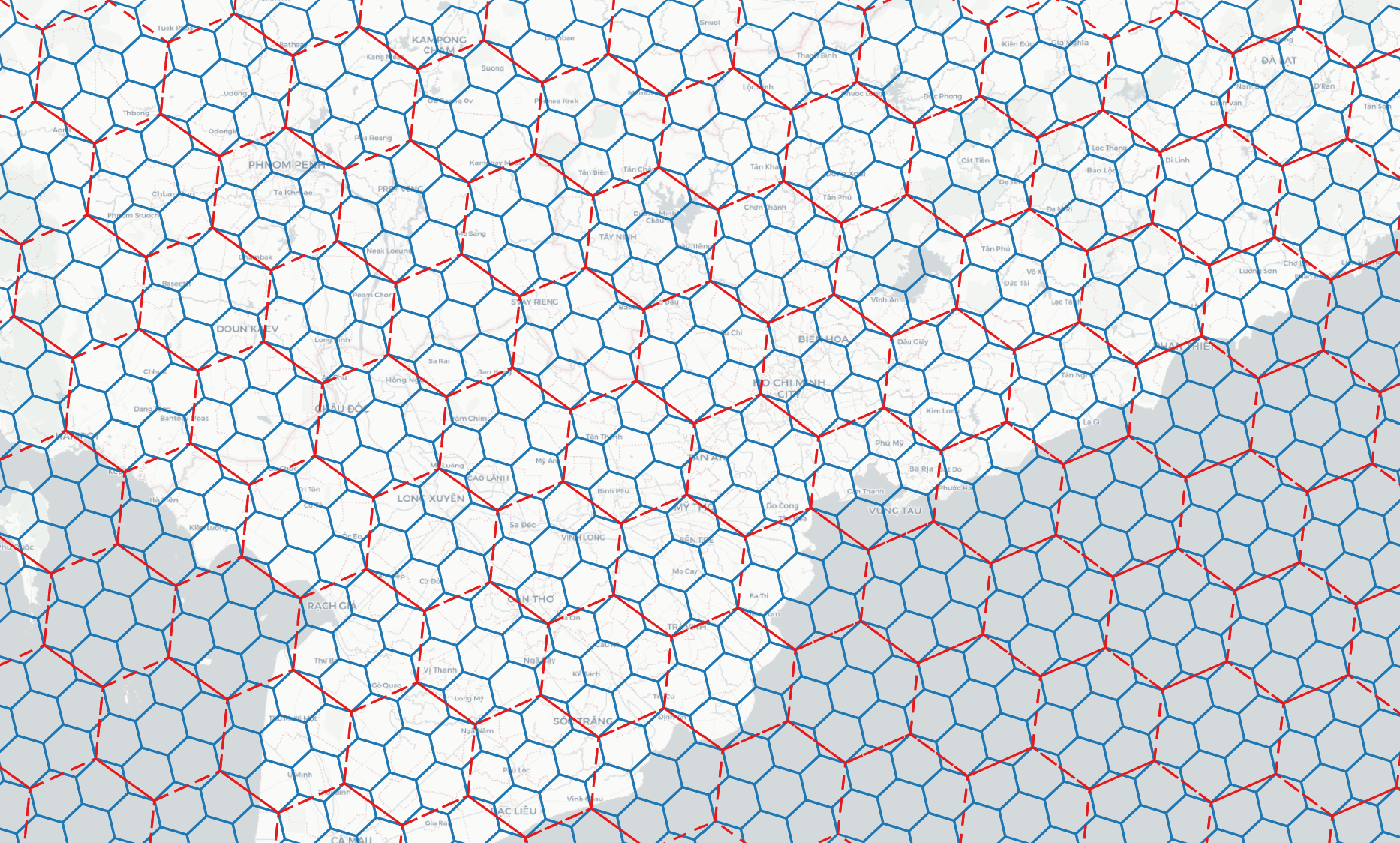
Generate DGGS at a specific resolution within a bounding box
> h3grid -r 11 -b 106.699007 10.762811 106.717674 10.778649 # h3grid -r <resolution> [0..15] -b <min_lon> <min_lat> <max_lon> <max_lat>
> s2grid -r 18 -b 106.699007 10.762811 106.717674 10.778649 # s2grid -r <resolution> [0..30] -b <min_lon> <min_lat> <max_lon> <max_lat>
> a5grid -r 18 -b 106.699007 10.762811 106.717674 10.778649 # a5grid -r <resolution> [0..29] -b <min_lon> <min_lat> <max_lon> <max_lat>
> rhealpixgrid -r 11 -b 106.699007 10.762811 106.717674 10.778649 # rhealpix2grid -r <resolution> [0..30] -b <min_lon> <min_lat> <max_lon> <max_lat>
> isea4tgrid -r 17 -b 106.699007 10.762811 106.717674 10.778649 # isea4tgrid -r <resolution> [0..25] -b <min_lon> <min_lat> <max_lon> <max_lat>
> isea3hgrid -r 20 -b 106.699007 10.762811 106.717674 10.778649 # isea3hgrid -r <resolution> [0..32] -b <min_lon> <min_lat> <max_lon> <max_lat>> isea3hstats # Number of cells, Avg Edge Length, Avg Cell Area at each resolution
> easegrid -r 4 -b 106.6990073571 10.7628112647 106.71767427 10.778649620 # easegrid -r <resolution> [0..6] -b <min_lon> <min_lat> <max_lon> <max_lat>
> dggridgen -t ISEA3H -r 2 -a ZORDER # Linux only: dggrid -t <DGGS Type> -r <resolution> -a<address_type>.
> qtmgrid -r 8 -b 106.6990073571 10.7628112647 106.71767427 10.778649620 # qtmgrid -r <resolution> [1..24] -b <min_lon> <min_lat> <max_lon> <max_lat>
> olcgrid -r 8 -b 106.6990073571 10.7628112647 106.71767427 10.778649620 # olcgrid -r <resolution> [2,4,6,8,10..15] -b <min_lon> <min_lat> <max_lon> <max_lat>
> geohashgrid -r 6 -b 106.699007 10.762811 106.717674 10.778649 # geohashgrid -r <resolution> [1..10] -b <min_lon> <min_lat> <max_lon> <max_lat> 1
> geohashgrid -r 2 -b 106.699007 10.762811 106.717674 10.778649 # geohashgrid -r <resolution> [0..5] -b <min_lon> <min_lat> <max_lon> <max_lat>
> mgrsgrid -r 1 -gzd 48P # geohashgrid -r <resolution> [0..5] -gzd <Grid Zone Designator, e.g. 48P>
> tilecodegrid -r 20 -b 106.699007 10.762811 106.717674 10.778649 # tilecodegrid -r <resolution> [0..26]
> quadkeygrid -r 20 -b 106.699007 10.762811 106.717674 10.778649 # quadkeygrid -r <resolution> [0..26]
> maidenheadgrid -r 4 -b 106.699007 10.762811 106.717674 10.778649 # maidenheadgrid -r <resolution> [1..4] -b <min_lon> <min_lat> <max_lon> <max_lat>
> garsgrid -r 1 -b 106.699007 10.762811 106.717674 10.778649 # garsgrid -r <resolution> [1..4] (30,15,5,1 minutes) -b <min_lon> <min_lat> <max_lon> <max_lat>
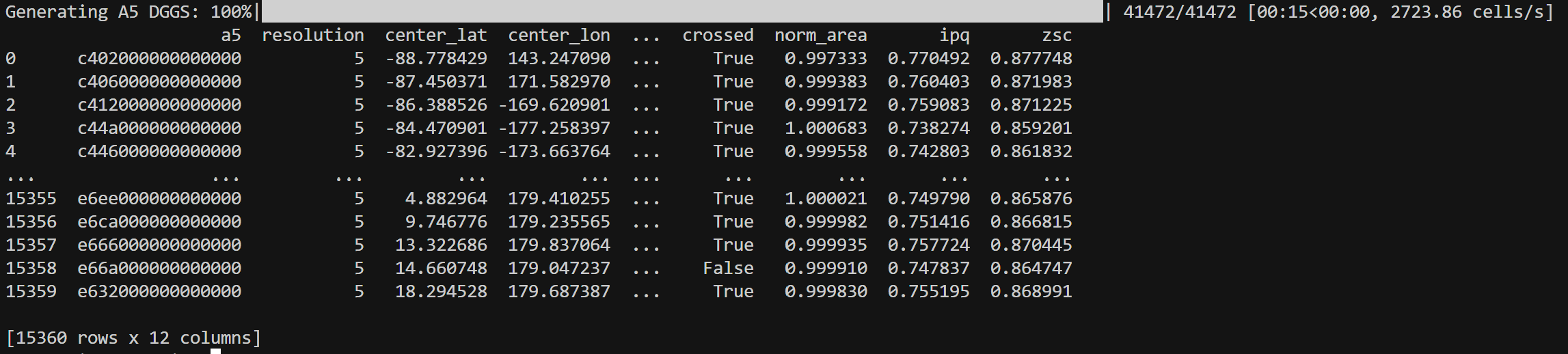
> h3inspect -r 3
> s2inspect -r 6
> a5inspect -r 5
> rhealpixinspect -r 4
> isea4tinspect -r 5
> isea3hinspect -r 3
> easeinspect -r 0
> qtminspect -r 6
> olcinspect -r 4
> geohashinspect -r 3
> georeftats -r 1
> tilecodeinspect -r 7
> quadkeyinspect -r 7
> maidenheadinspect -r 2
> garsinspect -r 1
> dggridinspect -t ISEA4T -r 3
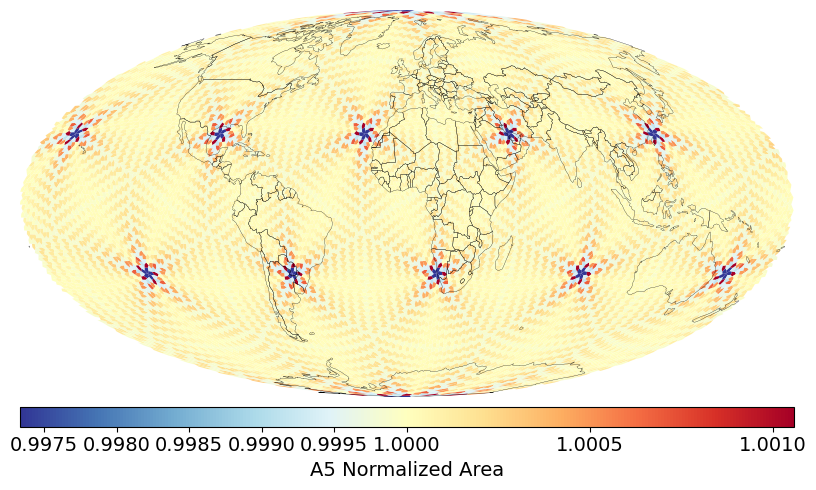
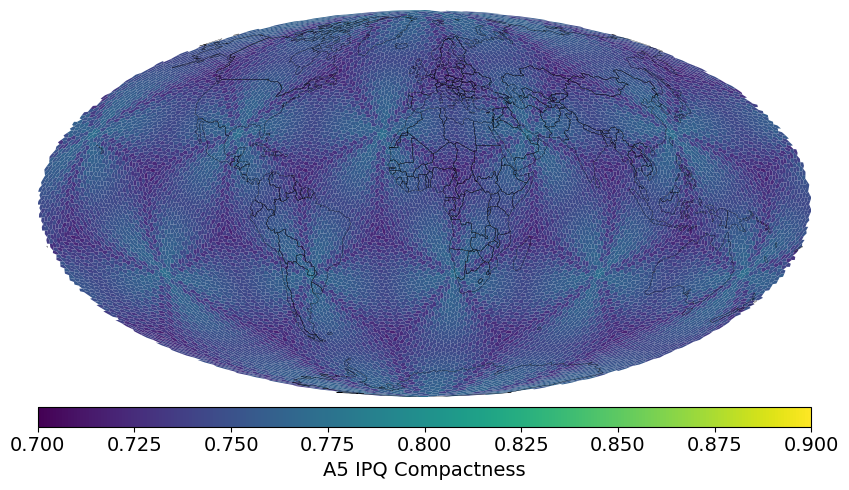
> h3stats # Number of cells, Avg Edge Length, Avg Cell Area at each resolution
> s2stats # Number of cells, Avg Edge Length, Avg Cell Area at each resolution
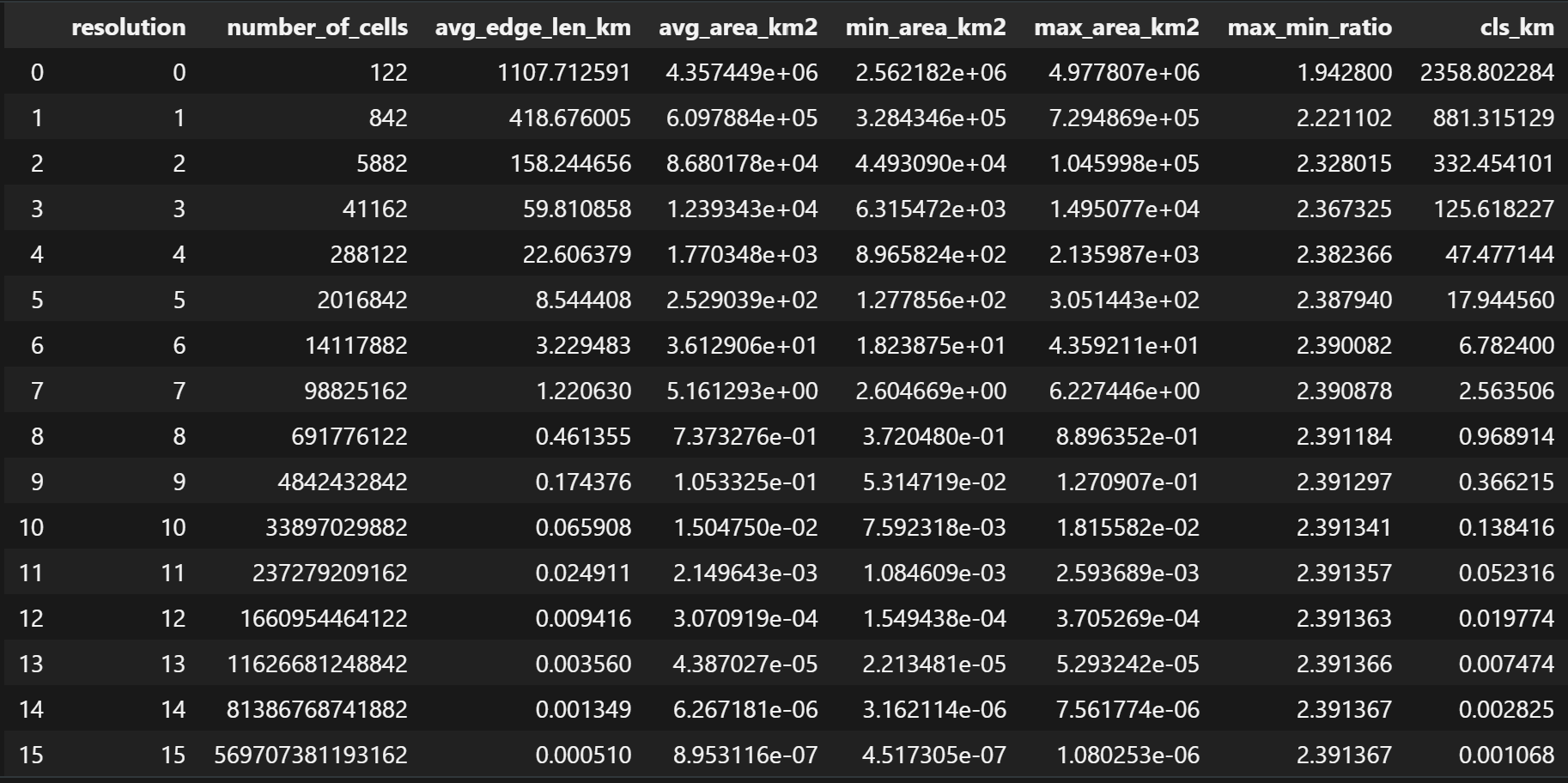
> a5stats # Number of cells, Avg Edge Length, Avg Cell Area at each resolution
> rhealpixstats # Number of cells, Avg Edge Length, Avg Cell Area at each resolution
> isea4tstats # Number of cells, Avg Edge Length, Avg Cell Area at each resolution
> isea3hstats # Number of cells, Avg Edge Length, Avg Cell Area at each resolution
> dggridstats -t FULLER3H -r 8 # Linux only: dggrid -t <DGGS Type> -r <resolution>
> easestats # Number of cells, Avg Edge Length, Avg Cell Area at each resolution
> qtmstats # Number of cells, Avg Edge Length, Avg Cell Area at each resolution
> olcstats # Number of cells, Avg Edge Length, Avg Cell Area at each resolution
> geohashstats # Number of cells, Avg Edge Length, Avg Cell Area at each resolution
> georeftats # Number of cells, Avg Edge Length, Avg Cell Area at each resolution
> mgrstats # Number of cells, Avg Edge Length, Avg Cell Area at each resolution
> tilecodestats # Number of cells, Cell Width, Cell Height, Cell Area at each resolution
> quadkeystats # Number of cells, Cell Width, Cell Height, Cell Area at each resolution
> maidenheadstats # Number of cells, Avg Edge Length, Avg Cell Area at each resolution
> garsstats # Number of cells, Avg Edge Length, Avg Cell Area at each resolution
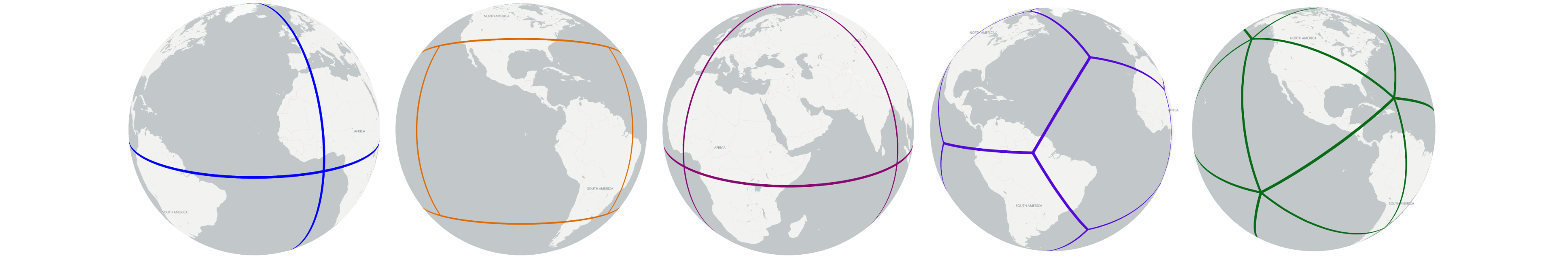
> tetrahedron # Generate Global Tetrahedron
> cube # Generate Global Cube
> octahedron # Generate Global Octahedron
> fuller_icosahedron # Generate Global Fuller Icosahedron
> rhombic_icosahedron # Generate Global rhombic Icosahedron
FAQs
Vgrid - DGGS and Cell-based Geocoding Utilites
We found that vgrid demonstrated a healthy version release cadence and project activity because the last version was released less than a year ago. It has 1 open source maintainer collaborating on the project.
Did you know?

Socket for GitHub automatically highlights issues in each pull request and monitors the health of all your open source dependencies. Discover the contents of your packages and block harmful activity before you install or update your dependencies.

Security News
OWASP’s 2025 Top 10 introduces Software Supply Chain Failures as a new category, reflecting rising concern over dependency and build system risks.
Research
/Security News
Socket researchers discovered nine malicious NuGet packages that use time-delayed payloads to crash applications and corrupt industrial control systems.

Security News
Socket CTO Ahmad Nassri discusses why supply chain attacks now target developer machines and what AI means for the future of enterprise security.