Product
Announcing Socket Fix 2.0
Socket Fix 2.0 brings targeted CVE remediation, smarter upgrade planning, and broader ecosystem support to help developers get to zero alerts.
connectome-interpreter
Advanced tools
This package is intended to be used for interpreting connectomics data.
To install:
pip install connectome-interpreter
Or to install the bleeding edge development version:
pip install git+https://github.com/YijieYin/connectome_interpreter.git
Documentation here (with some text snippets explaining various things).
Data obtained from Dorkenwald et al. 2024, Schlegel et al. 2024, and Matsliah et al. 2024. To visualise the neurons, you can use this url: https://tinyurl.com/flywire783. By using the connectivity information, you agree to follow the FlyWire citation guidelines and principles.
Data obtained from neuPrint and Nern et al. 2024, with the help of neuprint-python.
Data from Winding et al. 2023. You can also e.g. visualise the neurons in 3D in catmaid.
To facilitate neural circuit interpretation, we compile a list of cell types with known, experimentally tested, functions. This example notebook uses this list for query of neuron receptive field. The list aims to serve as a quick look-up of literature, instead of a stipulation of neural function.
; (semicolon + space), to facilitate programmatic access.Using connectome_interpreter, we compare the published connectomes against published experimental papers:
scipy.sparse.matrix (.npz) format for the adjacency matrices; and in .csv for the metadata.yy432atcam.ac.uk :).FAQs
A tool for connectomics data interpretation
We found that connectome-interpreter demonstrated a healthy version release cadence and project activity because the last version was released less than a year ago. It has 1 open source maintainer collaborating on the project.
Did you know?

Socket for GitHub automatically highlights issues in each pull request and monitors the health of all your open source dependencies. Discover the contents of your packages and block harmful activity before you install or update your dependencies.

Product
Socket Fix 2.0 brings targeted CVE remediation, smarter upgrade planning, and broader ecosystem support to help developers get to zero alerts.

Security News
Socket CEO Feross Aboukhadijeh joins Risky Business Weekly to unpack recent npm phishing attacks, their limited impact, and the risks if attackers get smarter.
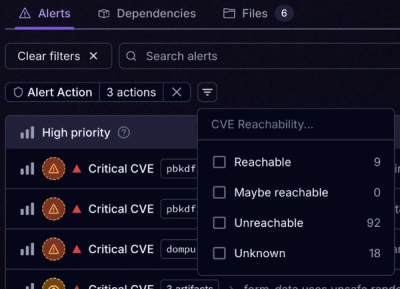
Product
Socket’s new Tier 1 Reachability filters out up to 80% of irrelevant CVEs, so security teams can focus on the vulnerabilities that matter.