Product
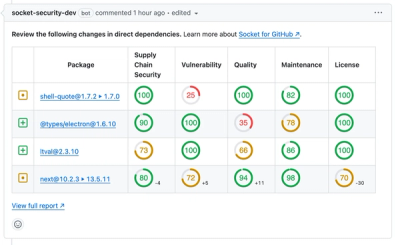
A New Design for GitHub PR Comments
We redesigned our GitHub PR comments to deliver clear, actionable security insights without adding noise to your workflow.
IPTK is a library specialized in the analysis of HLA-peptidomes identified through an immunopeptidomics pipeline.
IPTK is a Python library specialized in the analysis of HLA-peptidomes identified through an immunopeptidomic(IP) pipeline. The library provides a high level API for analyzing and visualizing the identified peptides, integrating transcritomics and protein structure information for a rich analysis of the identified immunopeptidomes. It also provides a toolbox for integrating and comparing different experiments and/or different runs.
Version 0.6 brings major upgrades to the library and introduce a wide array of function and classes for automating and accelerating IPTK performance
1- IPTK can now parse and work with mzIdentML files using the function parse_mzIdentML_to_identification_table define in the IO module of the library
2- IPTK can now process and read mzML files directly using PyOpenMS
3- IPTK has an improved function executional speed thanks to the AcceleratedFunctions module in the Analysis module which provides an acceleration using Numba
4- Current release also introduce, the Wrappers module which provide a simple abstraction for creating Experiment and ExperimentSet
5- Introducing ReplicatedExperiments which provides a simple API for creating experiments obtained from replicates
6- IPTK, current support concurrent execution, the wrapper submodules, now utilizes multiprocessing for parsing and reading multiple datasets on-parallel
7- Introducing, chordDiagram for showing overlap among experiments and Proband of experiments
1- Introducing GOEngine class which provides an easy-to-use wrapper around goatools for performing GOEA on the identified proteins.
2- Current release supports Jaccard index as a metric of similarity among experiments
3- Introducing support for visualizing GOEA results
4- Correction of minor bugs and documentation typos in previous releases
The major plan is to, first, increase and enhance IPTK scale and execution speed by offloading computational intensive tasks to RUST. Second, increase automation by providing custom analysis recipes for performing commonly used routines. Third, provide an API for integrating other omics layers, namely metabolomics and proteomics. Finally, adding support to PTM modified HLA peptides and proteins
1. Release 0.7.1 will aim at supporting the integration of Proteomic data with the library
2. Release 0.7.2 will aim at supporting the integration of Metabolomics data with the library
3. Release 0.7.3 will aim at standardizing all omics API and provide a high-level abstraction for working with them
4. Release 0.7.4-0.7.7 will aim at re-implement all the class in Rust and provide a python wrapper around these classes, Thus ensuring fast and concurrent execution
1. Release 0.8.1-0.8.4 will aim at re-implement all IPTK parsers in Rust and provide a python binder to it
2. Release 0.8.5-0.8.8 will aim at re-implementing all analysis function using Rust
Different minor releases will introduce different analysis Recipes to automate analysis tasks
IPTK version 1.0 is release on PyPi and on BioConda
1- Adding a class to query AFND database for allele frequency world-wide.
2- Adding function for plotting a choropleth for allele frequencies.
3- Adding classes for working directly with mzML files using pyopenMS framework
4- An experimental class that act as database interface and provide method for storing and querying immunopeptidomic data
Adding more control to the function plot_MDS_from_ic_coverage to fine-tune its behavior, for example, by controlling the random seed.
Corrected a bug in the Experiment class to correctly compute the length of peptides containing parentheses. This bug caused the len function to return the number of characters in the sequence instead of the number of amino acids.
Corrected a bug in the Peptide class to manage peptides containing parentheses in the sequence. This bug caused the len function to return the number of characters in the sequence instead of the number of amino acids.
Minor corrections in the visualization module
Minor corrections in the documentation and the default values for some parameters in the visualization functions
1- Adding function to compute immunopeptiomic coverage matrix
2- Introducing MDS plots for comparing the similarities between runs based on immunopeptidomic coverage
The library have three notebooks that provide a step-by-step guidance to use the library and to utilize its major APIs for interacting with an IPs data. These tutorial can be found at the Tutorial directory at the project's Github page.
IPTK has been documented using Sphinx, the manual of the library can be found at docs directory and online at readthedocs
The library can be installed using pip as follows:
pip install iptkl --user
The project was funded by the German Research Foundation (DFG) (Research Training Group 1743, ‘Genes, Environment and Inflammation’).
FAQs
IPTK is a library specialized in the analysis of HLA-peptidomes identified through an immunopeptidomics pipeline.
We found that iptkl demonstrated a healthy version release cadence and project activity because the last version was released less than a year ago. It has 1 open source maintainer collaborating on the project.
Did you know?

Socket for GitHub automatically highlights issues in each pull request and monitors the health of all your open source dependencies. Discover the contents of your packages and block harmful activity before you install or update your dependencies.

Product
We redesigned our GitHub PR comments to deliver clear, actionable security insights without adding noise to your workflow.
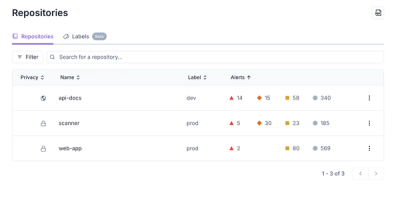
Product
Our redesigned Repositories page adds alert severity, filtering, and tabs for faster triage and clearer insights across all your projects.

Security News
Slopsquatting is a new supply chain threat where AI-assisted code generators recommend hallucinated packages that attackers register and weaponize.