Product
Announcing Socket Fix 2.0
Socket Fix 2.0 brings targeted CVE remediation, smarter upgrade planning, and broader ecosystem support to help developers get to zero alerts.
Whole Slide Image (WSI) conversion for brightfield histology images.
Note: This is in early development and there will likely be frequent and breaking changes.
Provides a command line interface (CLI) for easy convertion between formats:
Usage: wsic convert [OPTIONS]
Convert a WSI.
Options:
-i, --in-path PATH Path to WSI to read from.
-o, --out-path PATH The path to output to.
-t, --tile-size <INTEGER INTEGER>...
The size of the tiles to write.
-rt, --read-tile-size <INTEGER INTEGER>...
The size of the tiles to read.
-w, --workers INTEGER The number of workers to use.
-c, --compression [blosc|deflate|jpeg xl|jpeg-ls|jpeg|jpeg2000|lzw|png|webp|zstd]
The compression to use.
-cl, --compression-level INTEGER
The compression level to use.
-d, --downsample INTEGER The downsample factor to use.
-mpp, --microns-per-pixel <FLOAT FLOAT>...
The microns per pixel to use.
-ome, --ome / --no-ome Save with OME-TIFF metadata (OME-TIFF and
NGFF).
--overwrite / --no-overwrite Whether to overwrite the output file.
-to, --timeout FLOAT Timeout in seconds for reading a tile.
-W, --writer [auto|jp2|svs|tiff|zarr]
The writer to use. Overrides writer detected
by output file extension.
-s, --store [dir|ndir|zip|sqlite]
The store to use (zarr/NGFF only). Defaults
to ndir (nested directory).
-h, --help Show this message and exit.

For basic usage see the documentation page "How do I...?".
There are many other great tools in this space. Below are some other tools for converting WSIs.
Part of the Bio-Formats command line tools. Uses bioformats to convert from many formats to OME-TIFF.
https://www.openmicroscopy.org/bio-formats/downloads/
Convert from Bio-Formats formats to zarr.
https://github.com/glencoesoftware/bioformats2raw
Convert from Philips' iSyntax format to a zarr.
https://github.com/glencoesoftware/isyntax2raw
Convert OpenSlide images to WSI DICOM.
https://github.com/sectra-medical/wsidicomizer
This package was created with Cookiecutter and the audreyr/cookiecutter-pypackage project template.
-s argment for convert to specify store for zarr.DICOMWSIReader required user input at init.TIFFWriter pyramid building.DICOMWSIReader init warning and only warn from the main process.ZarrReaderWriter to seperate ZarrWriter and ZarrReader.CoordinateTransformation field default factor.get_tile method to all Reader classes.pyramid_downsamples argument when None.FAQs
Whole Slide Image (WSI) conversion for brightfield histology images
We found that wsic demonstrated a healthy version release cadence and project activity because the last version was released less than a year ago. It has 1 open source maintainer collaborating on the project.
Did you know?

Socket for GitHub automatically highlights issues in each pull request and monitors the health of all your open source dependencies. Discover the contents of your packages and block harmful activity before you install or update your dependencies.

Product
Socket Fix 2.0 brings targeted CVE remediation, smarter upgrade planning, and broader ecosystem support to help developers get to zero alerts.

Security News
Socket CEO Feross Aboukhadijeh joins Risky Business Weekly to unpack recent npm phishing attacks, their limited impact, and the risks if attackers get smarter.
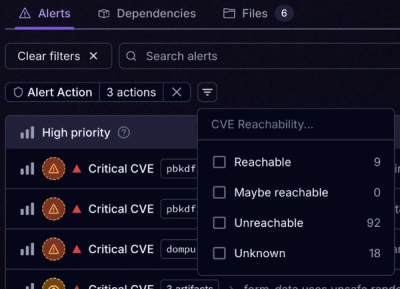
Product
Socket’s new Tier 1 Reachability filters out up to 80% of irrelevant CVEs, so security teams can focus on the vulnerabilities that matter.