Robust comparative analysis and contamination removal for metagenomics

With Recentrifuge, researchers can interactively explore what organisms are in their samples and at which level of confidence, thus enabling a robust comparative analysis of multiple samples in any metagenomic study.
- Removes diverse contaminants, including crossovers, using a novel robust contamination removal algorithm.
- Provides a confidence level for every result, since the calculated score propagates to all the downstream analysis and comparisons.
- Unveils the generalities and specificities in the metagenomic samples, thanks to a new comparative analysis engine.
Recentrifuge's novel approach combines robust statistics, arithmetic of scored taxonomic trees, and parallel computational algorithms.
Recentrifuge is especially useful when a more reliable detection of minority organisms is needed (e.g. in the case of low microbial biomass metagenomic studies) in clinical, environmental, or forensic analysis. Beyond the standard confidence levels, Recentrifuge implements others devoted to variable length reads, very convenient for complex datasets generated by nanopore sequencers.
To play with an example of a webpage generated by Recentrifuge, click on the next screenshot:
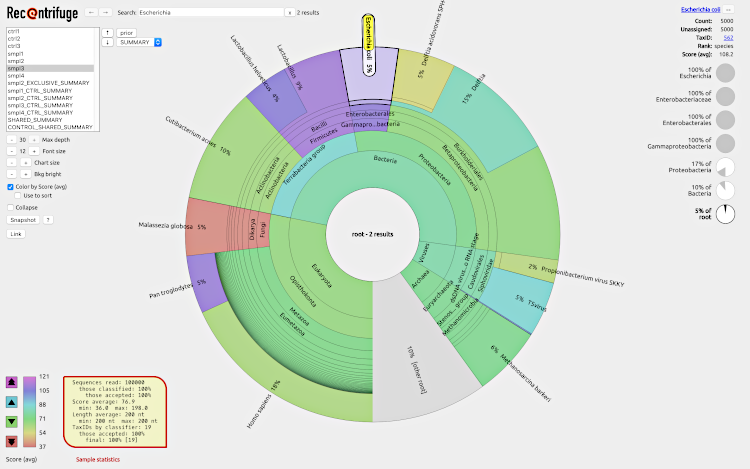
Recentrifuge webpage example