
Security News
Risky Biz Podcast: Making Reachability Analysis Work in Real-World Codebases
This episode explores the hard problem of reachability analysis, from static analysis limits to handling dynamic languages and massive dependency trees.
The Ruby UCSC API: accessing the UCSC Genome Database using Ruby.
Your comments, suggestions and requests are welcome. Documentation and feedback are available at the UserEcho site at http://rubyucscapi.userecho.com/.
The Ruby UCSC API: accessing the UCSC genome database using Ruby: Hiroyuki Mishima, Jan Aerts, Toshiaki Katayama, Raoul JP Bonnal and Koh-ichiro Yoshiura, BMC Bioinformatics 13:240 (2012).
doi:10.1186/1471-2105-13-240
http://www.biomedcentral.com/1471-2105/13/240/
$ gem install bio-ucsc-api
TogoWS ( http://togows.org/ ), a web-service of DBCLS ( http://dbcls.rois.ac.jp/ ), supports UCSC Genome Database and offers the REST interface using the Ruby UCSC API internally. Please see the TogoWS API documentation (the "External API" section) at http://togows.org/help/ .
If your favorite databese is not supported, please do not hesitate to contact the author because the author mainly working only with human genomes.
This package is based on the followings:
Supported Ruby interpreter implementations:
Ruby Version 2.0.0 or later
Ruby version 1.9.3 or later
JRuby version 1.6.3 or later - Appropiate Java heap size may have to be specified to invoke JRuby, especially when you use Bio::Ucsc::File::Twobit. Try "jruby -J-Xmx3g your_script.rb" to keep 3G byte heap.
Ruby version 1.8.7 or earlier are no longer supported by UCSC API v0.6.0 and later because Ruby on Rails and ActiveRecord version 4.0 do not support these old Rubies.
Major rubygem dependencies:
ActiveSupport::Deprecation.silenced = true.See also:
ActiveSupport::Deprecation.silenced = true. Because ActiveRecord v4.0 does not support Ruby v1.8.7 and earlier, Ruby UCSC API no longer supports these older Rubies.See 'ChangeLog.md' for older changes.
Bio::Ucsc module. For example, the human hg19 database is referred by Bio::Ucsc::Hg19.Bio::Ucsc::Hg19.connect.Bio::Ucsc::Hg19::Snp138.Bio::Ucsc::Hg19::Snp138.first.chr1:1233-5678. If a table to query has the "bin" column, the bin index system is automatically used to speed-up the query.result.name.At first, you have to declare the API and establish the connection to a database.
require 'bio-ucsc'
DB = Bio::Ucsc::Hg19
DB.connect
# Suppressing deprecation warnings for using dynamic finders such as 'find_by_name_and_chrom'.
# These syles are deprecated in ActiveRecord 4.0.
ActiveSupport::Deprecation.silenced = true
Table search using genomic intervals:
require 'bio-ucsc'
DB = Bio::Ucsc::Hg19
DB.connect
DB::Snp138.with_interval("chr1:1-11,000").find(:all).each do |e|
i = GenomicInterval.zero_based(e.chrom, e.chromStart, e.chromEnd)
puts "#{i.chrom}\t#{i.chr_start}\t#{e.name}\t#{e[:class]}" # "e.class" does not work
end
gi = "chr17:7,579,614-7,579,700"
puts DB::Snp138.with_interval(gi).find(:all)
puts DB::Snp138.with_interval_excl(gi).find(:all)
relation = DB::Snp138.with_interval(gi).select(:name)
puts relation.to_sql
# => SELECT name FROM `snp138`
# WHERE (chrom = 'chr17' AND bin in (642,80,9,1,0)
# AND ((chromStart BETWEEN 7579613 AND 7579700) AND
# (chromEnd BETWEEN 7579613 AND 7579700)))
puts relation.find_all_by_class_and_strand("in-del", "+").size # => 1
# Rails4 style
puts DB::Snp138.where(name: "rs56289060").first
# Old style
ActiveSupport::Deprecation.silenced = true # Suppress warnings
puts DB::Snp138.find_all_by_name("rs56289060").first
Sometimes, queries using raw SQLs provide elegant solutions.
require 'bio-ucsc'
DB = Bio::Ucsc::Hg19
DB.connect
sql << 'SQL'
SELECT name,chrom,chromStart,chromEnd,observed
FROM snp138
WHERE name="rs56289060"
SQL
puts DB::Snp138.find_by_sql(sql)
For gene prediction (genePred) tables, such as RefSeq, EndGene, and WgEncodeGencodeBasicV12, Ruby UCSC API automatically implements #exon, #introns, #cdss (or an alias #cdses) methods. Exons, introns, and CDSes are accessible as Array objects of Bio::GenomicInterval.
require 'bio-ucsc'
DB = Bio::Ucsc::Hg19
DB.connect
r = DB::RefGene.with_interval("chr1:1,000,000-1,100,000").first
puts "gene strand = #{r.strand}"
r.exons.each{|x|puts "[#{x.chr_start}, #{x.chr_end}]"}
r.cdss.each{|x|puts "[#{x.chr_start}, #{x.chr_end}]"}
r.introns.each{|x|puts "[#{x.chr_start}, #{x.chr_end}]"}
retrieve reference sequence from a locally-stored 2bit file. The "hg19.2bit" file can be downloaded from http://hgdownload.cse.ucsc.edu/goldenPath/hg19/bigZips/hg19.2bit
hg19ref = Bio::Ucsc::File::Twobit.load("hg19.2bit")
puts hg19ref.find_by_interval("chr1:9,500-10,999")
# another way to access a twobit file
puts Bio::Ucsc::File::Twobit.open("hg19.2bit"){|tb|tb.find_by_interval("chr1:9,500-10,999")}
Connetcting to non-official or local full/partial mirror MySQL servers
Bio::Ucsc::Hg18.connect( :db_host => 'localhost',
:db_username => 'genome',
:db_password => '' )
Bio::Ucsc::Hg18.default # reset to connect UCSC's public MySQL sever
Bio::Ucsc::Hg18.connect
And see also sample scripts in the samples directory.
T2micron_est class.HInv and NIAGene, respectively.find_by_interval or .find_all_by_interval. These methods look for correspondent separated tables automatically. However, you cannot combine with other find_by_[field] methods. Moreover, if you have to perform single- or multi-chromosomal search, you have to access separated tables individually and integrate results by yourself. Fortunately, recent databases, including hg19, seem not to use chromosome-specific tables.attribute, valid, validate, class, method, methods, and type cannot be accessed using instance methods. This restriction is because of the collision of method names that are internally used by ActiveRecord. Instead, use hash to access the field like result[:type].Ruby UCSC API supports two ways to define table association/relation, manual and automatic. Manual definition can define minimum association set you need. Automatic definition is easy to use. However, automatic definition may define huge number of association. You may have to restrict database set before definition.
See samples/snp2gene.rb. Association definitions using has_one/has_many methods are shown below. class_eval is used not to replace but to add definition.
Bio::Ucsc::Hg19::KnownGene.class_eval do
has_one :knownToEnsembl, {:primary_key => :name, :foreign_key => :name}
end
Bio::Ucsc::Hg19::KnownToEnsembl.class_eval do
has_one :ensGtp, {:primary_key => :value, :foreign_key => :transcript}
has_one :kgXref, {:primary_key => :name, :foreign_key => :kgID}
end
Bio::Ucsc::Hg19::KgXref.class_eval do
has_one :refLink, {:primary_key => :mRNA, :foreign_key => :mrnaAcc}
end
And fields can be referred like the followings:
kg.knownToEnsembl.ensGtp.gene
kg.knownToEnsembl.kgXref.geneSymbol
kg.knownToEnsembl.kgXref.refLink.mrnaAcc
ActiveRecord::Base#find can be used with the :include option to perform "eager fetching" to reduce number of SQL statement submission.
kg = Bio::Ucsc::Hg19::KnownGene.with_interval(gi).
find(:first,
:include => [:knownToEnsembl => :ensGtp,
:knownToEnsembl => {:kgXref => :refLink}])
First, use Bio::Ucsc::Joiner.load(url) to the all.joiner file from url. If url is not given, http://genome-source.cse.ucsc.edu/gitweb/?p=kent.git;a=blob_plain;f=src/hg/makeDb/schema/all.joiner;hb=HEAD will be used as the url. Please see further infomation about all.joiner at http://genome-source.cse.ucsc.edu/gitweb/?p=kent.git;a=blob;f=src/hg/makeDb/schema/joiner.doc;hb=HEAD
Next, you can overwrite all.joiner variables by the Joiner#variables method. For examle, a "gbd" variable means "all databases". Overwriting this variables can restrict databases to be used in table assocations and makes automatic definition faster. Unconnected databases and undefined tables are ignored during definition.
Then, you can access an associated tables using a method. Note that automatic definition always use "has_many" methods. Thus, resuls are always returned as an array.
Bio::Ucsc::Hg19.connect
Bio::Ucsc::Hg18.connect
joiner = Bio::Ucsc::Schema::Joiner.load
joiner.variables["gbd"] = ["hg19", "hg18"]
joiner.define_association(Bio::Ucsc::Hg19::Snp138)
# "first" is required because the snp138Seq method always returns an array.
puts Bio::Ucsc::Hg19::Snp138.find_by_name("rs242").snp138Seq.first.file_offset
Copyright: (c) 2011-2018 MISHIMA, Hiroyuki (hmishima at nagasaki-u.ac.jp / Twitter: @mishima_eng (in English) and @mishimahryk (in Japanese)
Copyright: (c) 2010 Jan Aerts
License: The MIT license. See LICENSE.txt for further details..
FAQs
Unknown package
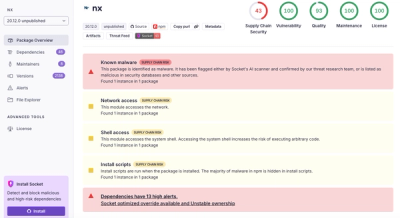
We found that bio-ucsc-api demonstrated a not healthy version release cadence and project activity because the last version was released a year ago. It has 2 open source maintainers collaborating on the project.
Did you know?

Socket for GitHub automatically highlights issues in each pull request and monitors the health of all your open source dependencies. Discover the contents of your packages and block harmful activity before you install or update your dependencies.

Security News
This episode explores the hard problem of reachability analysis, from static analysis limits to handling dynamic languages and massive dependency trees.
Security News
/Research
Malicious Nx npm versions stole secrets and wallet info using AI CLI tools; Socket’s AI scanner detected the supply chain attack and flagged the malware.

Security News
CISA’s 2025 draft SBOM guidance adds new fields like hashes, licenses, and tool metadata to make software inventories more actionable.