Security News
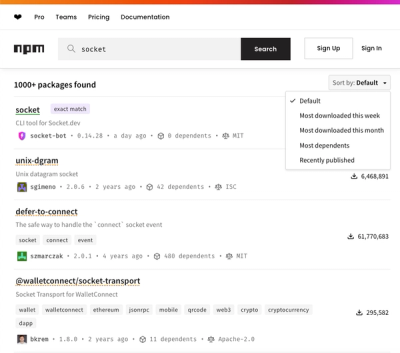
npm Updates Search Experience with New Objective Sorting Options
npm has a revamped search experience with new, more transparent sorting options—Relevance, Downloads, Dependents, and Publish Date.
@jbrowse/cli
Advanced tools
This document covers the CLI tools. Note: for @jbrowse/img static export tool, see https://www.npmjs.com/package/@jbrowse/img
The command line tools can be installed globally using npm as follows
$ npm install -g @jbrowse/cli
A CLI tool called jbrowse should then be available in the path. You can test
your installation with
$ jbrowse --version
It is also possible to do one-off executions using npx, e.g.
npx @jbrowse/cli create myfolder
It is likely preferable in most cases to install the tools globally with npm install @jbrowse/cli -g however
jbrowse add-assembly SEQUENCEjbrowse add-connection CONNECTIONURLORPATHjbrowse add-track TRACKjbrowse add-track-json TRACKjbrowse admin-serverjbrowse create LOCALPATHjbrowse help [COMMAND]jbrowse set-default-sessionjbrowse text-indexjbrowse upgrade [LOCALPATH]jbrowse add-assembly SEQUENCEAdd an assembly to a JBrowse 2 configuration
USAGE
$ jbrowse add-assembly SEQUENCE
ARGUMENTS
SEQUENCE
sequence file or URL
If TYPE is indexedFasta or bgzipFasta, the index file defaults to <location>.fai
and can be optionally specified with --faiLocation
If TYPE is bgzipFasta, the gzip index file defaults to <location>.gzi and can be
optionally specified with --gziLocation
OPTIONS
-a, --alias=alias
An alias for the assembly name (e.g. "hg38" if the name of the assembly is "GRCh38");
can be specified multiple times
-f, --force
Equivalent to `--skipCheck --overwrite`
-h, --help
show CLI help
-l, --load=copy|symlink|move|inPlace
Required flag when using a local file. Choose how to manage the data directory. Copy, symlink, or move the data
directory to the JBrowse directory. Or use inPlace to modify the config without doing any file operations
-n, --name=name
Name of the assembly; if not specified, will be guessed using the sequence file name
-t, --type=indexedFasta|bgzipFasta|twoBit|chromSizes|custom
type of sequence, by default inferred from sequence file
indexedFasta An index FASTA (e.g. .fa or .fasta) file;
can optionally specify --faiLocation
bgzipFasta A block-gzipped and indexed FASTA (e.g. .fa.gz or .fasta.gz) file;
can optionally specify --faiLocation and/or --gziLocation
twoBit A twoBit (e.g. .2bit) file
chromSizes A chromosome sizes (e.g. .chrom.sizes) file
custom Either a JSON file location or inline JSON that defines a custom
sequence adapter; must provide --name if using inline JSON
--displayName=displayName
The display name to specify for the assembly, e.g. "Homo sapiens (hg38)" while the name can be a shorter identifier
like "hg38"
--faiLocation=faiLocation
[default: <fastaLocation>.fai] FASTA index file or URL
--gziLocation=gziLocation
[default: <fastaLocation>.gzi] FASTA gzip index file or URL
--out=out
synonym for target
--overwrite
Overwrite existing assembly if one with the same name exists
--refNameAliases=refNameAliases
Reference sequence name aliases file or URL; assumed to be a tab-separated aliases
file unless --refNameAliasesType is specified
--refNameAliasesType=aliases|custom
Type of aliases defined by --refNameAliases; if "custom", --refNameAliases is either
a JSON file location or inline JSON that defines a custom sequence adapter
--refNameColors=refNameColors
A comma-separated list of color strings for the reference sequence names; will cycle
through colors if there are fewer colors than sequences
--skipCheck
Don't check whether or not the sequence file or URL exists or if you are in a JBrowse directory
--target=target
path to config file in JB2 installation directory to write out to.
Creates ./config.json if nonexistent
EXAMPLES
# add assembly to installation in current directory. assumes .fai file also exists, and copies GRCh38.fa and
GRCh38.fa.fai to current directory
$ jbrowse add-assembly GRCh38.fa --load copy
# add assembly to a specific jb2 installation path using --out, and copies the .fa and .fa.fai file to /path/to/jb2
$ jbrowse add-assembly GRCh38.fa --out /path/to/jb2/ --load copy
# force indexedFasta for add-assembly without relying on file extension
$ jbrowse add-assembly GRCh38.xyz --type indexedFasta --load copy
# add displayName for an assembly
$ jbrowse add-assembly myFile.fa.gz --name hg38 --displayName "Homo sapiens (hg38)"
# use chrom.sizes file for assembly instead of a fasta file
$ jbrowse add-assembly GRCh38.chrom.sizes --load inPlace
# add assembly from preconfigured json file, expert option
$ jbrowse add-assembly GRCh38.config.json --load copy
# add assembly from a 2bit file, also note pointing direct to a URL so no --load flag needed
$ jbrowse add-assembly https://example.com/data/sample.2bit
# add a bgzip indexed fasta inferred by fa.gz extension. assumes .fa.gz.gzi and .fa.gz.fai files also exists
$ jbrowse add-assembly myfile.fa.gz --load copy
See code: src/commands/add-assembly.ts
jbrowse add-connection CONNECTIONURLORPATHAdd a connection to a JBrowse 2 configuration
USAGE
$ jbrowse add-connection CONNECTIONURLORPATH
ARGUMENTS
CONNECTIONURLORPATH URL of data directory
For hub file, usually called hub.txt
For JBrowse 1, location of JB1 data directory similar to http://mysite.com/jbrowse/data/
OPTIONS
-a, --assemblyName=assemblyName Assembly name of the connection If none, will default to the assembly in your config
file
-c, --config=config Any extra config settings to add to connection in JSON object format, such as
'{"uri":"url":"https://sample.com"}, "locationType": "UriLocation"}'
-f, --force Equivalent to `--skipCheck --overwrite`
-h, --help show CLI help
-n, --name=name Name of the connection. Defaults to connectionId if not provided
-t, --type=type type of connection, ex. JBrowse1Connection, UCSCTrackHubConnection, custom
--connectionId=connectionId Id for the connection that must be unique to JBrowse. Defaults to
'connectionType-assemblyName-currentTime'
--out=out synonym for target
--overwrite Overwrites any existing connections if same connection id
--skipCheck Don't check whether or not the data directory URL exists or if you are in a JBrowse
directory
--target=target path to config file in JB2 installation directory to write out to.
EXAMPLES
$ jbrowse add-connection http://mysite.com/jbrowse/data/
$ jbrowse add-connection http://mysite.com/jbrowse/custom_data_folder/ --type JBrowse1Connection
$ jbrowse add-connection http://mysite.com/path/to/hub.txt --assemblyName hg19
$ jbrowse add-connection http://mysite.com/path/to/custom_hub_name.txt --type UCSCTrackHubConnection --assemblyName
hg19
$ jbrowse add-connection http://mysite.com/path/to/custom --type custom --config
'{"uri":{"url":"https://mysite.com/path/to/custom"}, "locationType": "UriLocation"}' --assemblyName hg19
$ jbrowse add-connection https://mysite.com/path/to/hub.txt --connectionId newId --name newName --target
/path/to/jb2/installation/config.json
See code: src/commands/add-connection.ts
jbrowse add-track TRACKAdd a track to a JBrowse 2 configuration
USAGE
$ jbrowse add-track TRACK
ARGUMENTS
TRACK Track file or URL
OPTIONS
-a, --assemblyNames=assemblyNames Assembly name or names for track as comma separated string. If none, will
default to the assembly in your config file
-d, --description=description Optional description of the track
-f, --force Equivalent to `--skipCheck --overwrite`
-h, --help show CLI help
-l, --load=copy|symlink|move|inPlace Required flag when using a local file. Choose how to manage the track. Copy,
symlink, or move the track to the JBrowse directory. Or inPlace to leave track
alone
-n, --name=name Name of the track. Will be defaulted to the trackId if none specified
-t, --trackType=trackType Type of track, by default inferred from track file
--category=category Optional Comma separated string of categories to group tracks
--config=config Any extra config settings to add to a track. i.e '{"defaultRendering":
"density"}'
--indexFile=indexFile Optional index file for the track
--out=out synonym for target
--overwrite Overwrites existing track if it shares the same trackId
--protocol=protocol [default: uri] Force protocol to a specific value
--skipCheck Skip check for whether or not the file or URL exists or if you are in a JBrowse
directory
--subDir=subDir when using --load a file, output to a subdirectory of the target dir
--target=target path to config file in JB2 installation to write out to.
--trackId=trackId trackId for the track, by default inferred from filename, must be unique
throughout config
EXAMPLES
# copy /path/to/my.bam and /path/to/my.bam.bai to current directory and adds track to config.json
$ jbrowse add-track /path/to/my.bam --load copy
# copy my.bam and my.bam.bai to /path/to/jb2/bam and adds track entry to /path/to/jb2/bam/config.json
$ jbrowse add-track my.bam --load copy --out /path/to/jb2 --subDir bam
# same as above, but specify path to bai file. needed for if the bai file does not have the extension .bam.bai
$ jbrowse add-track my.bam --indexFile my.bai --load copy
# creates symlink for /path/to/my.bam and adds track to config.json
$ jbrowse add-track /path/to/my.bam --load symlink
# add track from URL to config.json, no --load flag needed
$ jbrowse add-track https://mywebsite.com/my.bam
# --load inPlace adds a track without doing file operations
$ jbrowse add-track /url/relative/path.bam --load inPlace
See code: src/commands/add-track.ts
jbrowse add-track-json TRACKAdd a track configuration directly from a JSON hunk to the JBrowse 2 configuration
USAGE
$ jbrowse add-track-json TRACK
ARGUMENTS
TRACK track JSON file or command line arg blob
OPTIONS
-u, --update update the contents of an existing track, matched based on trackId
--out=out synonym for target
--target=target path to config file in JB2 installation directory to write out to.
Creates ./config.json if nonexistent
EXAMPLES
$ jbrowse add-track-json track.json
$ jbrowse add-track-json track.json --update
See code: src/commands/add-track-json.ts
jbrowse admin-serverStart up a small admin server for JBrowse configuration
USAGE
$ jbrowse admin-server
OPTIONS
-h, --help show CLI help
-p, --port=port Specifified port to start the server on;
Default is 9090.
--out=out synonym for target
--target=target path to config file in JB2 installation directory to write out to.
Creates ./config.json if nonexistent
EXAMPLES
$ jbrowse admin-server
$ jbrowse admin-server -p 8888
See code: src/commands/admin-server.ts
jbrowse create LOCALPATHDownloads and installs the latest JBrowse 2 release
USAGE
$ jbrowse create LOCALPATH
ARGUMENTS
LOCALPATH Location where JBrowse 2 will be installed
OPTIONS
-f, --force Overwrites existing JBrowse 2 installation if present in path
-h, --help show CLI help
-l, --listVersions Lists out all versions of JBrowse 2
-t, --tag=tag Version of JBrowse 2 to install. Format is v1.0.0.
Defaults to latest
-u, --url=url A direct URL to a JBrowse 2 release
--branch=branch Download a development build from a named git branch
--nightly Download the latest development build from the main branch
EXAMPLES
# Download latest release from github, and put in specific path
$ jbrowse create /path/to/new/installation
# Download latest release from github and force overwrite existing contents at path
$ jbrowse create /path/to/new/installation --force
# Download latest release from a specific URL
$ jbrowse create /path/to/new/installation --url url.com/directjbrowselink.zip
# Download a specific tag from github
$ jbrowse create /path/to/new/installation --tag v1.0.0
# List available versions
$ jbrowse create --listVersions
See code: src/commands/create.ts
jbrowse help [COMMAND]display help for jbrowse
USAGE
$ jbrowse help [COMMAND]
ARGUMENTS
COMMAND command to show help for
OPTIONS
--all see all commands in CLI
See code: @oclif/plugin-help
jbrowse set-default-sessionSet a default session with views and tracks
USAGE
$ jbrowse set-default-session
OPTIONS
-c, --currentSession List out the current default session
-h, --help show CLI help
-n, --name=name [default: New Default Session] Give a name for the default session
-s, --session=session set path to a file containing session in json format
-t, --tracks=tracks Track id or track ids as comma separated string to put into default session
-v, --view=view View type in config to be added as default session, i.e LinearGenomeView, CircularView,
DotplotView.
Must be provided if no default session file provided
--delete Delete any existing default session.
--out=out synonym for target
--target=target path to config file in JB2 installation directory to write out to
--viewId=viewId Identifier for the view. Will be generated on default
EXAMPLES
$ jbrowse set-default-session --session /path/to/default/session.json
$ jbrowse set-default-session --target /path/to/jb2/installation/config.json --view LinearGenomeView --tracks track1,
track2, track3
$ jbrowse set-default-session --view LinearGenomeView, --name newName --viewId view-no-tracks
$ jbrowse set-default-session --currentSession # Prints out current default session
See code: src/commands/set-default-session.ts
jbrowse text-indexMake a text-indexing file for any given track(s).
USAGE
$ jbrowse text-index
OPTIONS
-a, --assemblies=assemblies Specify the assembl(ies) to create an index for. If unspecified, creates an index for
each assembly in the config
-h, --help show CLI help
-q, --quiet Hide the progress bars
--attributes=attributes [default: Name,ID] Comma separated list of attributes to index
--dryrun Just print out tracks that will be indexed by the process, without doing any indexing
--exclude=exclude [default: CDS,exon] Adds gene type to list of excluded types
--file=file File or files to index (can be used to create trix indexes for embedded component use
cases not using a config.json for example)
--force Overwrite previously existing indexes
--out=out Synonym for target
--perTrack If set, creates an index per track
--target=target Path to config file in JB2 installation directory to read from.
--tracks=tracks Specific tracks to index, formatted as comma separated trackIds. If unspecified, indexes
all available tracks
EXAMPLES
# indexes all tracks that it can find in the current directory's config.json
$ jbrowse text-index
# indexes specific trackIds that it can find in the current directory's config.json
$ jbrowse text-index --tracks=track1,track2,track3
# indexes all tracks in a directory's config.json or in a specific config file
$ jbrowse text-index --out /path/to/jb2/
# indexes only a specific assembly, and overwrite what was previously there using force (which is needed if a previous
index already existed)
$ jbrowse text-index -a hg19 --force
# create index for some files for use in @jbrowse/react-linear-genome-view or similar
$ jbrowse text-index --file myfile.gff3.gz --file myfile.vcfgz --out indexes
See code: src/commands/text-index.ts
jbrowse upgrade [LOCALPATH]Upgrades JBrowse 2 to latest version
USAGE
$ jbrowse upgrade [LOCALPATH]
ARGUMENTS
LOCALPATH [default: .] Location where JBrowse 2 is installed
OPTIONS
-h, --help show CLI help
-l, --listVersions Lists out all versions of JBrowse 2
-t, --tag=tag Version of JBrowse 2 to install. Format is v1.0.0.
Defaults to latest
-u, --url=url A direct URL to a JBrowse 2 release
--branch=branch Download a development build from a named git branch
--nightly Download the latest development build from the main branch
EXAMPLES
# Upgrades current directory to latest jbrowse release
$ jbrowse upgrade
# Upgrade jbrowse instance at a specific filesystem path
$ jbrowse upgrade /path/to/jbrowse2/installation
# Upgrade to a specific tag
$ jbrowse upgrade /path/to/jbrowse2/installation --tag v1.0.0
# List versions available on github
$ jbrowse upgrade --listVersions
# Upgrade from a specific URL
$ jbrowse upgrade --url https://sample.com/jbrowse2.zip
# Get nightly release from main branch
$ jbrowse upgrade --nightly
See code: src/commands/upgrade.ts
Debug logs (provided by debug) can be
printed by setting the DEBUG environment variable. Setting DEBUG=* will
print all debug logs. Setting DEBUG=jbrowse* will print only logs from this
tool, and setting e.g. DEBUG=jbrowse:add-assembly will print only logs from
the add-assembly command.
1.5.4 (2022-01-07)
<details><summary>Packages in this release</summary> <p>| Package | Download | | --------------------------------------- | ----------------------------------------------------------------- | | @jbrowse/core | https://www.npmjs.com/package/@jbrowse/core | | @jbrowse/development-tools | https://www.npmjs.com/package/@jbrowse/development-tools | | @jbrowse/plugin-alignments | https://www.npmjs.com/package/@jbrowse/plugin-alignments | | @jbrowse/plugin-authentication | | | @jbrowse/plugin-bed | https://www.npmjs.com/package/@jbrowse/plugin-bed | | @jbrowse/plugin-breakpoint-split-view | | | @jbrowse/plugin-circular-view | https://www.npmjs.com/package/@jbrowse/plugin-circular-view | | @jbrowse/plugin-config | https://www.npmjs.com/package/@jbrowse/plugin-config | | @jbrowse/plugin-data-management | https://www.npmjs.com/package/@jbrowse/plugin-data-management | | @jbrowse/plugin-dotplot-view | | | @jbrowse/plugin-gff3 | https://www.npmjs.com/package/@jbrowse/plugin-gff3 | | @jbrowse/plugin-grid-bookmark | https://www.npmjs.com/package/@jbrowse/plugin-grid-bookmark | | @jbrowse/plugin-gtf | https://www.npmjs.com/package/@jbrowse/plugin-gtf | | @jbrowse/plugin-hic | | | @jbrowse/plugin-legacy-jbrowse | https://www.npmjs.com/package/@jbrowse/plugin-legacy-jbrowse | | @jbrowse/plugin-linear-comparative-view | | | @jbrowse/plugin-linear-genome-view | https://www.npmjs.com/package/@jbrowse/plugin-linear-genome-view | | @jbrowse/plugin-sequence | https://www.npmjs.com/package/@jbrowse/plugin-sequence | | @jbrowse/plugin-spreadsheet-view | | | @jbrowse/plugin-sv-inspector | | | @jbrowse/plugin-svg | https://www.npmjs.com/package/@jbrowse/plugin-svg | | @jbrowse/plugin-variants | https://www.npmjs.com/package/@jbrowse/plugin-variants | | @jbrowse/plugin-wiggle | https://www.npmjs.com/package/@jbrowse/plugin-wiggle | | @jbrowse/cli | https://www.npmjs.com/package/@jbrowse/cli | | @jbrowse/desktop | | | @jbrowse/img | https://www.npmjs.com/package/@jbrowse/img | | @jbrowse/react-circular-genome-view | https://www.npmjs.com/package/@jbrowse/react-circular-genome-view | | @jbrowse/react-linear-genome-view | https://www.npmjs.com/package/@jbrowse/react-linear-genome-view | | @jbrowse/web | |
</p> </details>core
core
core
FAQs
A command line tool for working with JBrowse 2
The npm package @jbrowse/cli receives a total of 180 weekly downloads. As such, @jbrowse/cli popularity was classified as not popular.
We found that @jbrowse/cli demonstrated a healthy version release cadence and project activity because the last version was released less than a year ago. It has 0 open source maintainers collaborating on the project.
Did you know?

Socket for GitHub automatically highlights issues in each pull request and monitors the health of all your open source dependencies. Discover the contents of your packages and block harmful activity before you install or update your dependencies.

Security News
npm has a revamped search experience with new, more transparent sorting options—Relevance, Downloads, Dependents, and Publish Date.

Security News
A supply chain attack has been detected in versions 1.95.6 and 1.95.7 of the popular @solana/web3.js library.
Research
Security News
A malicious npm package targets Solana developers, rerouting funds in 2% of transactions to a hardcoded address.