Security News
Create React App Officially Deprecated Amid React 19 Compatibility Issues
Create React App is officially deprecated due to React 19 issues and lack of maintenance—developers should switch to Vite or other modern alternatives.
constellation-js
Advanced tools
Constellation is a tool for parsing a language specifying genetic parts and their relationships
git clone git@github.com:hicsail/constellation-js.git
npm run build && npm run start
Then open http://localhost:8082/ on browser
The package can be installed in the following way.
npm install constellation-js
It is possible to generate a collection of designs that match a specification using graph construction and traversals.
const constellation = require('constellation-js');
let goldbar = '{PT7_a then galK}';
let categories = {
"PT7_a":{
"promoter": [
"PT7_WTa",
"PT7_3a",
"PT7_1a"
]
},
"galK":{
"cds": [
"galK"
]
}
}
let result = constellation.goldbar(goldbar, categories, {designName: 'my-first-design'});
// result.stateGraph, result.designs, result.sbol
| Optional parameters | Description |
|---|---|
designName | Name of design space for SBOL output, defaults is "constellation-design" |
numDesigns | Max number of designs to enumerate, default is 20 |
maxCycles | Cycle depth for -orMore operators, default is 0 |
representation | Choose between EDGE or NODE based graph, default is EDGE |
andTolerance | Choose between 0, 1, 2 for the AND operator, default is 0 |
mergeTolerance | Choose between 0, 1, 2 for the MERGE operator, default is 0 |
| Output | Description |
|---|---|
stateGraph | See Graph Data Structure |
designs | List of enumerated designs |
sbol | See Synthetic Biology Open Language |
It is also possible to generate a collection of designs that match a specification using a purely symbolic approach (note that this approach supports only a tolerance of 0 for the AND operator and does not support the MERGE operator).
let result = constellation.symbolic(
"(one-or-more a) then (one-or-more x)",
{"a": {"b": ["c"]}, "x":{"y": ["z1", "z2", "z3"]}},
{"numDesigns": 'all', "maxCycles":7});
GOLDBAR syntax for the case studies described in the manuscript are available here and can be demoed on Constellation's UI via the drop down menu.
Genetic design spaces in Constellation are represented in three ways:
The supported GOLDBAR concrete syntax for genetic design spaces is presented below using extended BNF notation. Notice that then and . are equivalent, and the delimiters (...) and {...} are equivalent.
<seq> ::= <exp> then <seq>
| <exp> . <seq>
| <exp>
<exp> ::= <term> or <exp>
| <term> and <exp>
| <term> merge <exp>
| <term>
<term> ::= one-or-more <term>
| zero-or-more <term>
| zero-or-one <term>
| reverse-comp <term>
| ( <seq> )
| { <seq> }
| <atom>
<atom> ::= ([A-Za-z0-9]|-|_)+
The JSON schema for the GOLDBAR abstract syntax tree representation (parsed from the concrete syntax presented above) can be found in schemas/ast.schema.json.
Constellation supports both NODE and EDGE based versions of a design space. Below are examples equivalent to the GOLDBAR specification promoter then one-or-more CDS then terminator
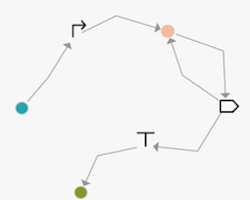 | 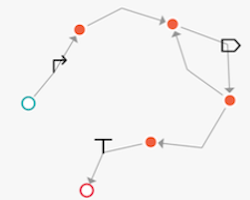 |
|---|---|
| Visualization of node-based graph | Visualization of edge-based graph |
The JSON schema for a design space graph can be found in schemas/graph.schema.json. Below is an example of a node-based graph within a single node in JSON format.
{
"604571a7-9e38-4647-afd0-9546399480b5": {
"id": "604571a7-9e38-4647-afd0-9546399480b5",
"text": "root",
"type": "root",
"edges": [
"b79407eb-95f0-4b78-99da-779f2c9cad46",
"7f6ca2fb-ef67-4687-924c-4285de896877"
],
"operator": ["One"]
}
}
SBOL is an open standard for the representation of in silico biological designs, and the icons used in this tool are provided by SBOL Visual. Design spaces are expressed in SBOL via the CombinatorialDerivation extension and can be exported and stored in Knox. This third form of design space representation allows Constellation to be easily integrated in the synthetic biology community.
FAQs
Constellation is a tool for parsing a language specifying genetic parts and their relationships
The npm package constellation-js receives a total of 3 weekly downloads. As such, constellation-js popularity was classified as not popular.
We found that constellation-js demonstrated a not healthy version release cadence and project activity because the last version was released a year ago. It has 1 open source maintainer collaborating on the project.
Did you know?

Socket for GitHub automatically highlights issues in each pull request and monitors the health of all your open source dependencies. Discover the contents of your packages and block harmful activity before you install or update your dependencies.

Security News
Create React App is officially deprecated due to React 19 issues and lack of maintenance—developers should switch to Vite or other modern alternatives.

Security News
Oracle seeks to dismiss fraud claims in the JavaScript trademark dispute, delaying the case and avoiding questions about its right to the name.

Security News
The Linux Foundation is warning open source developers that compliance with global sanctions is mandatory, highlighting legal risks and restrictions on contributions.