
Security News
Fluent Assertions Faces Backlash After Abandoning Open Source Licensing
Fluent Assertions is facing backlash after dropping the Apache license for a commercial model, leaving users blindsided and questioning contributor rights.
A fast and covariate-adaptive method for multiple hypothesis testing.
Software accompanying the paper "AdaFDR: a Fast, Powerful and Covariate-Adaptive Approach to Multiple Hypothesis Testing", 2018.
pip install adafdr
adafdr.method contains all methods while adafdr.data_loader contains the data.
They can be imported as
import adafdr.method as md
import adafdr.data_loader as dl
Other ways of importing are usually compatible. For example, one can import the package with import adafdr
and call method xxx in the method module via adafdr.method.xxx()
For a set of N hypotheses, the input data includes the p-values p and the d-dimensional covariate x,
with the following format:
p: (N,) numpy.ndarray.x: (N,d) numpy.ndarray.When d=1, x is allowed to be either (N,) numpy.ndarray
or (N,1) numpy.ndarray.
The covariate visualization method adafdr_explore can be used as
adafdr.method.adafdr_explore(p, x, output_folder=None, covariate_type=None)
If the output_folder is a filepath (str) instead of None, the covariate visualization figures will be
saved in output_folder. Otherwise, they will show up in the console.
covariate_type: a length-d python list with values 0/1. It specifies the type of each covariate: 0 means numerical/ordinal while 1 means categorical. For example, covariate_type=[0,1] means there are 2 covariates, the first is numerical/ordinal and the second is categorical. If not specified, a covariate with more than 75 distinct values is regarded as numerical/ordinal and otherwise categorical.
See also doc for more details.
The multiple hypothesis testing method adafdr_test can be used as
res = adafdr.method.adafdr_test(p, x, alpha=0.1, covariate_type=None)
res = adafdr.method.adafdr_test(p, x, alpha=0.1, fast_mode=False, covariate_type=None)
res = adafdr.method.adafdr_test(p, x, alpha=0.1, fast_mode=False, single_core=False, covariate_type=None)
res is a dictionary containing the results, including:
res['decision']: a (N,) boolean vector, decision for each hypothesis with value 1 meaning rejection.res['threshold']: a (N,) float vector, threshold for each hypothesis.If the output_folder is a filepath (str) instead of None, the logfiles and some intermediate results will be
saved in output_folder. Otherwise, they will show up in the console.
covariate_type: a length-d python list with values 0/1. It specifies the type of each covariate: 0 means numerical/ordinal while 1 means categorical. For example, covariate_type=[0,1] means there are 2 covariates, the first is numerical/ordinal and the second is categorical. If not specified, a covariate with more than 75 distinct values is regarded as numerical/ordinal and otherwise categorical.
See also doc for more details.
The following is an example on the airway RNA-seq data used in the paper.
Here we load the airway data used in the paper. See vignettes for other data accompanied with the package.
import adafdr.method as md
import adafdr.data_loader as dl
p,x = dl.data_airway()
adafdr_exploremd.adafdr_explore(p, x, output_folder=None)
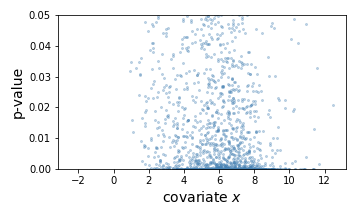
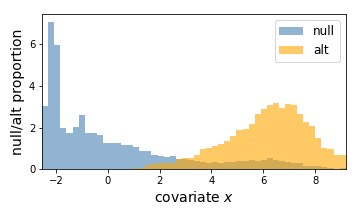
Here, the left is a scatter plot of each hypothesis with p-values (y-axis) plotted against the covariate (x-axis). The right panel shows the estimated null hypothesis distribution (blue) and the estimated alternative hypothesis distribution (orange) with respect to the covariate. Here we can conclude that a hypothesis is more likely to be significant if the covariate (gene expression) value is higher.
adafdr_testres = md.adafdr_test(p, x, fast_mode=True, output_folder=None)
Here, the learned threshold res['threshold'] looks as follows.
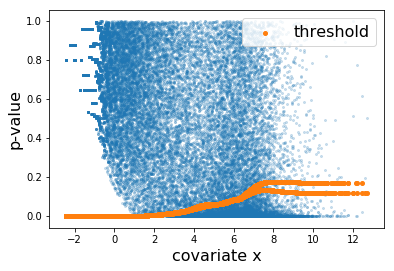
Each orange dot corresponds to the threhsold to one hypothesis. The discrepancy at the right is due to the difference between the thresholds learned by the two folds.
Here is a quick test. First check if the package can be successfully imported:
import adafdr.method as md
import adafdr.data_loader as dl
Next, run a small example which should take a few seconds:
import numpy as np
p,x,h,_,_ = dl.load_1d_bump_slope()
res = md.adafdr_test(p, x, alpha=0.1)
t = res['threshold']
D = np.sum(p<=t)
FD = np.sum((p<=t)&(~h))
print('# AdaFDR successfully finished!')
print('# D=%d, FD=%d, FDP=%0.3f'%(D, FD, FD/D))
It runs AdaFDR-fast on a 1d simulated data. If the package is successfully imported, the result should look like:
# AdaFDR successfully finished!
# D=837, FD=79, FDP=0.094
R API of this package can be found here.
Zhang, Martin J., Fei Xia, and James Zou. "AdaFDR: a Fast, Powerful and Covariate-Adaptive Approach to Multiple Hypothesis Testing." bioRxiv (2018): 496372.
Xia, Fei, et al. "Neuralfdr: Learning discovery thresholds from hypothesis features." Advances in Neural Information Processing Systems. 2017.
FAQs
A fast and covariate-adaptive method for multiple hypothesis testing
We found that adafdr demonstrated a healthy version release cadence and project activity because the last version was released less than a year ago. It has 1 open source maintainer collaborating on the project.
Did you know?

Socket for GitHub automatically highlights issues in each pull request and monitors the health of all your open source dependencies. Discover the contents of your packages and block harmful activity before you install or update your dependencies.

Security News
Fluent Assertions is facing backlash after dropping the Apache license for a commercial model, leaving users blindsided and questioning contributor rights.

Research
Security News
Socket researchers uncover the risks of a malicious Python package targeting Discord developers.

Security News
The UK is proposing a bold ban on ransomware payments by public entities to disrupt cybercrime, protect critical services, and lead global cybersecurity efforts.