pyGCO: a python wrapper for the graph cuts
The original wrapper is pygco







This is a python wrapper for gco-v3.0 package, which implements a graph cuts based move-making algorithm for optimization in Markov Random Fields.
It contains a copy of the gco-v3.0 package. Some of the design were borrowed from the gco_python package. However, compared to gco_python:
- This package does not depend on Cython. Instead it is implemented using the ctypes library and a C wrapper of the C++ code.
- This package is an almost complete wrapper for gco-v3.0, which supports more direct low level control over GCoptimization objects.
- This package supports graphs with edges weighted differently.
This wrapper is composed of two parts, a C wrapper and a python wrapper.
Implemented functions
- cut_general_graph(...)
- cut_grid_graph(...)
- cut_grid_graph_simple(...)
Building wrapper
- download the last version of gco-v3.0 to the gco_source
- compile gco-v3.0 and the C wrapper using
make - compile test_wrapper using
make test_wrapper - run the C test code
./test_wrapper (now you have the C wrapper ready)
make download
make all
make test_wrapper
./test_wrapper
The successful run should return:
labels = [ 0 2 2 1 ], energy=19
data energy=15, smooth energy=4
Next test the python wrapper using python test_examples.py, if it works fine you are ready to use pygco.
To include pygco in your code, simply import pygco module. See the documentation inside code for more details.
Install wrapper
Clone repository and enter folder, then
pip install -r requirements.txt
python setup.py install
Now it can be also installed from PyPi
pip install gco-wrapper
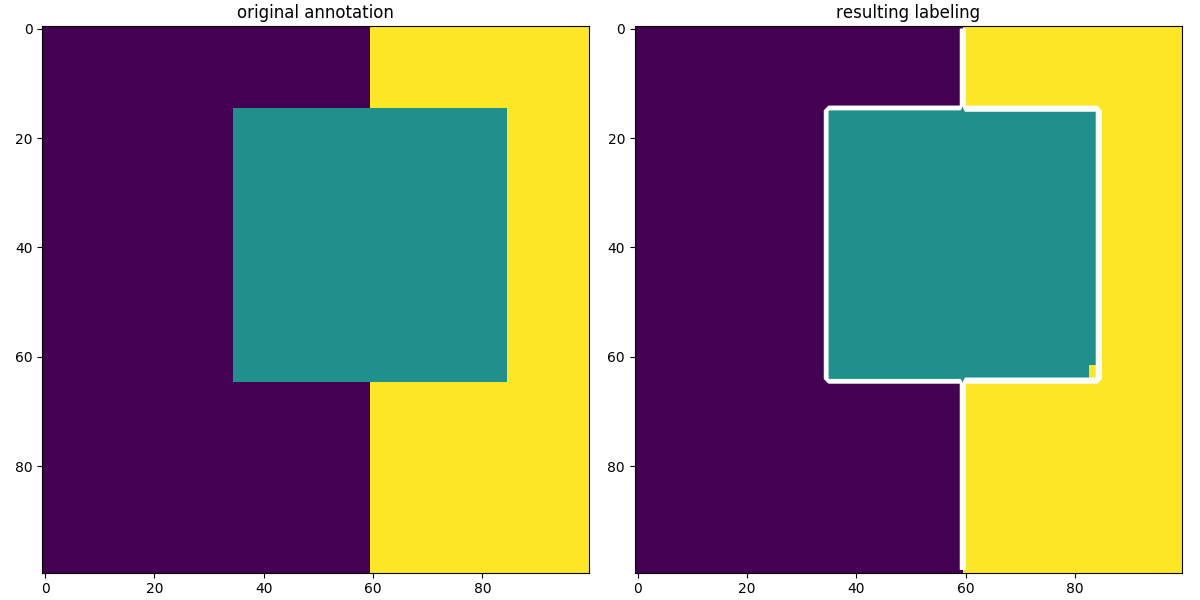
Show test results
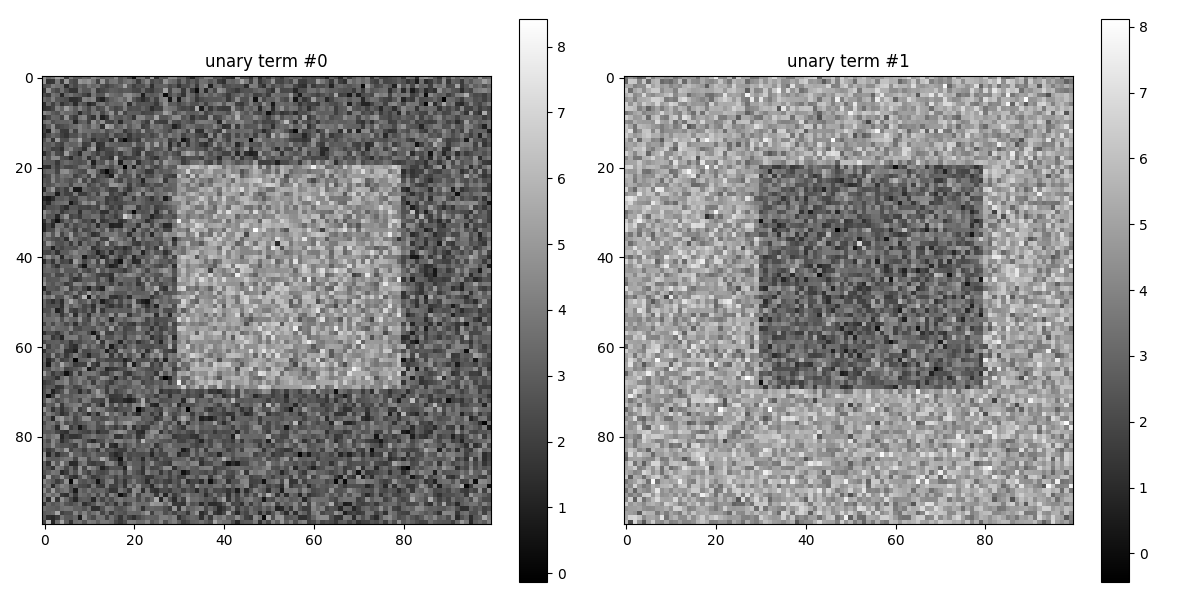
Visualisation of the unary terns for binary segmentation
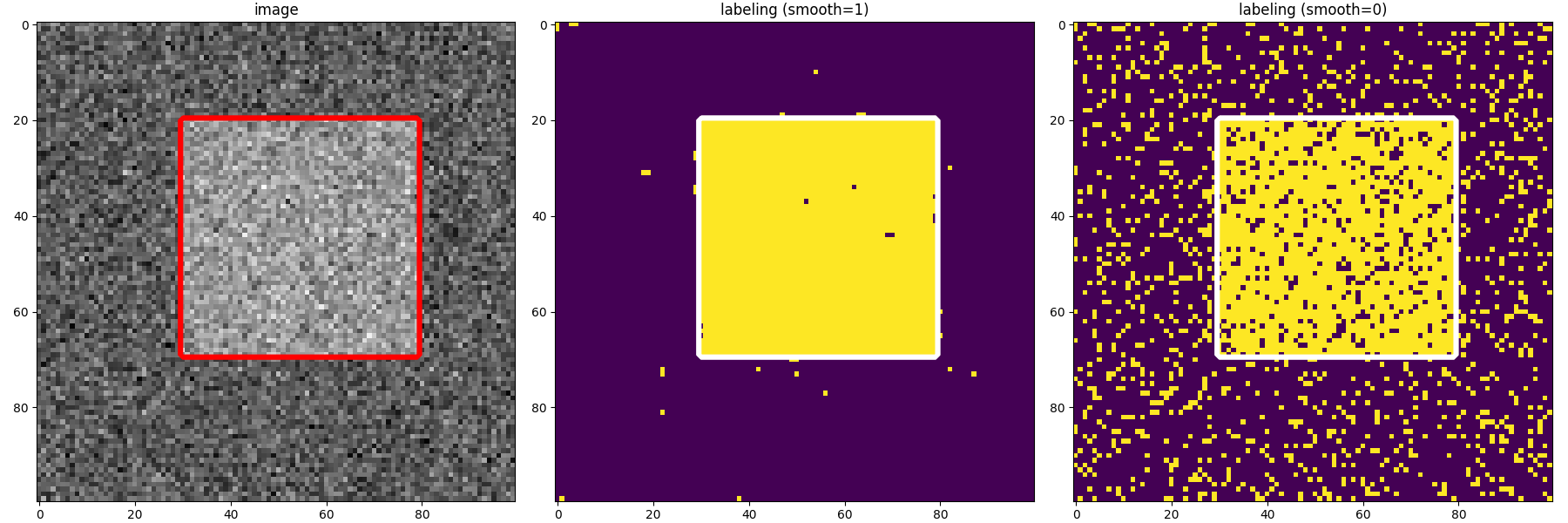
4-connected components with the initial labeling (left) and estimated labeling with regularisation 1 (middle) and 0 (right)
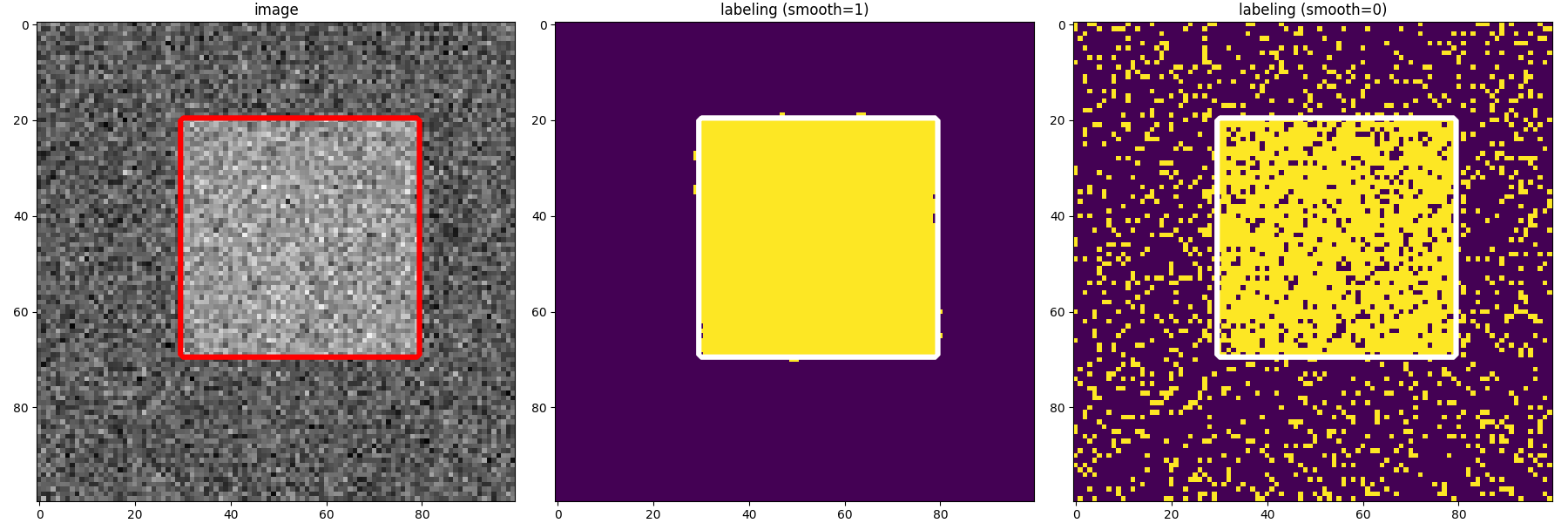
8-connected components with the initial labeling (left) and estimated labeling with regularisation 1 (middle) and 0 (right)
Visualisation of the unary terns for 3 labels segmentation
with the initial labeling (left) and estimated labeling (right)