






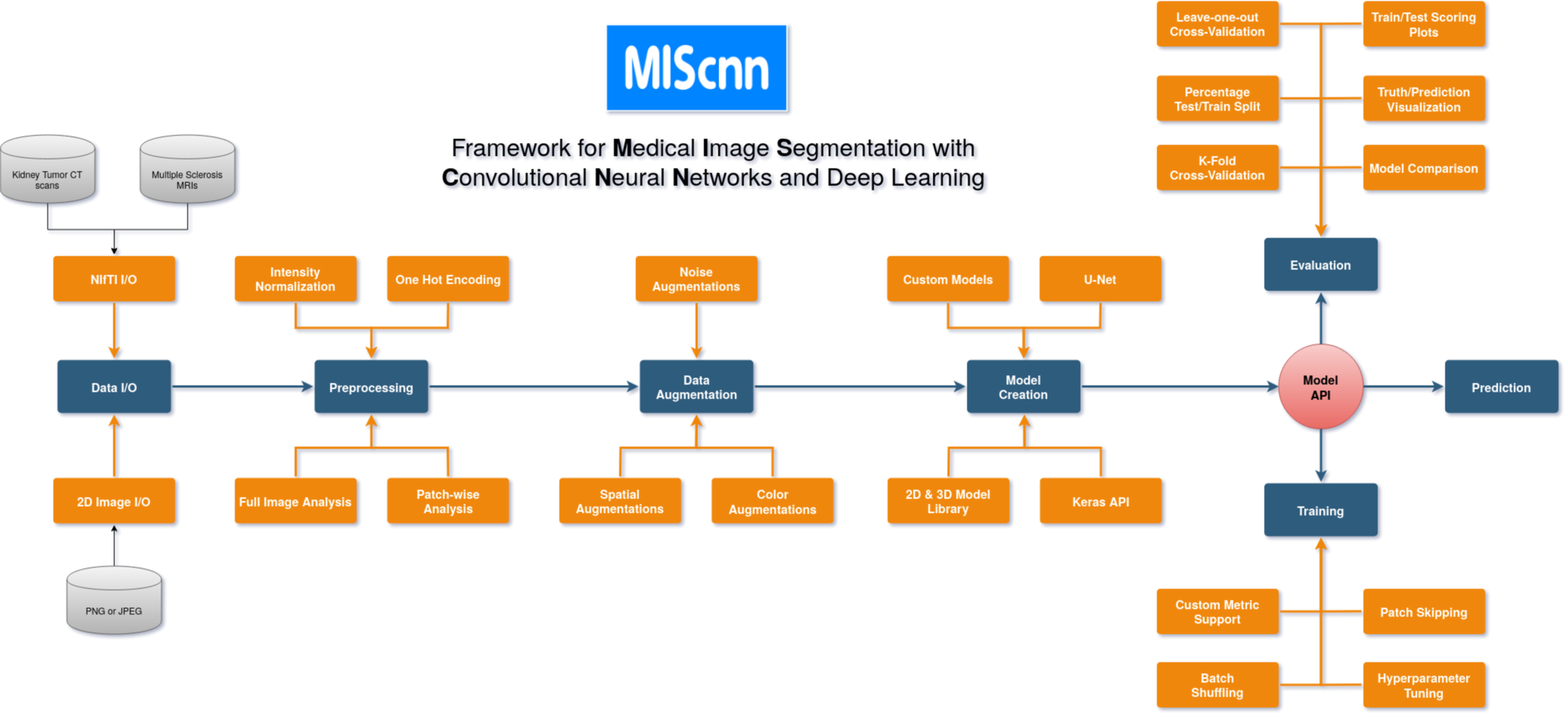
The open-source Python library MIScnn is an intuitive API allowing fast setup of medical image segmentation pipelines with state-of-the-art convolutional neural network and deep learning models in just a few lines of code.
MIScnn provides several core features:
- 2D/3D medical image segmentation for binary and multi-class problems
- Data I/O, preprocessing and data augmentation for biomedical images
- Patch-wise and full image analysis
- State-of-the-art deep learning model and metric library
- Intuitive and fast model utilization (training, prediction)
- Multiple automatic evaluation techniques (e.g. cross-validation)
- Custom model, data I/O, pre-/postprocessing and metric support
- Based on Keras with Tensorflow as backend
Resources
Author
Dominik Müller
Email: dominik.mueller@informatik.uni-augsburg.de
IT-Infrastructure for Translational Medical Research
University Augsburg
Augsburg, Bavaria, Germany
How to cite / More information
Dominik Müller and Frank Kramer. (2019)
MIScnn: A Framework for Medical Image Segmentation with Convolutional Neural Networks and Deep Learning.
arXiv e-print: https://arxiv.org/abs/1910.09308
Article{miscnn,
title={MIScnn: A Framework for Medical Image Segmentation with Convolutional Neural Networks and Deep Learning},
author={Dominik Müller and Frank Kramer},
year={2019},
eprint={1910.09308},
archivePrefix={arXiv},
primaryClass={eess.IV}
}
Thank you for citing our work.
License
This project is licensed under the GNU GENERAL PUBLIC LICENSE Version 3.
See the LICENSE.md file for license rights and limitations.











