
Research
Security News
Quasar RAT Disguised as an npm Package for Detecting Vulnerabilities in Ethereum Smart Contracts
Socket researchers uncover a malicious npm package posing as a tool for detecting vulnerabilities in Etherium smart contracts.
Fast, flexible, and advanced augmentation library for deep learning, computer vision, and medical imaging. Albumentations offers a wide range of transformations for both 2D (images, masks, bboxes, keypoints) and 3D (volumes, volumetric masks) data, with optimized performance and seamless integration into ML workflows.
Docs | Discord | Twitter | LinkedIn
Albumentations is a Python library for image augmentation. Image augmentation is used in deep learning and computer vision tasks to increase the quality of trained models. The purpose of image augmentation is to create new training samples from the existing data.
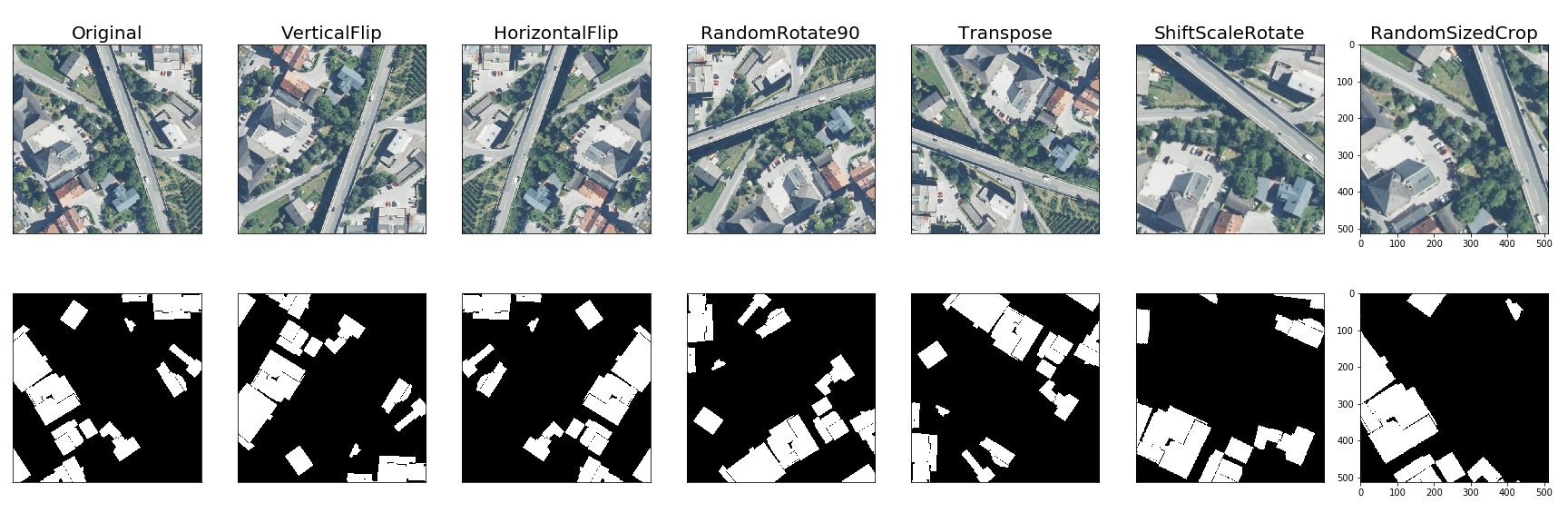
Here is an example of how you can apply some pixel-level augmentations from Albumentations to create new images from the original one:

Albumentations thrives on developer contributions. We appreciate our sponsors who help sustain the project's infrastructure.
| 🏆 Gold Sponsors |
|---|
| Your company could be here |
| 🥈 Silver Sponsors |
|---|
 |
| 🥉 Bronze Sponsors |
|---|
 |
Your sponsorship is a way to say "thank you" to the maintainers and contributors who spend their free time building and maintaining Albumentations. Sponsors are featured on our website and README. View sponsorship tiers on GitHub Sponsors
Vladimir I. Iglovikov | Kaggle Grandmaster
Mikhail Druzhinin | Kaggle Expert
Alexander Buslaev | Kaggle Master
Eugene Khvedchenya | Kaggle Grandmaster
Albumentations requires Python 3.9 or higher. To install the latest version from PyPI:
pip install -U albumentations
Other installation options are described in the documentation.
The full documentation is available at https://albumentations.ai/docs/.
import albumentations as A
import cv2
# Declare an augmentation pipeline
transform = A.Compose([
A.RandomCrop(width=256, height=256),
A.HorizontalFlip(p=0.5),
A.RandomBrightnessContrast(p=0.2),
])
# Read an image with OpenCV and convert it to the RGB colorspace
image = cv2.imread("image.jpg")
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
# Augment an image
transformed = transform(image=image)
transformed_image = transformed["image"]
Please start with the introduction articles about why image augmentation is important and how it helps to build better models.
If you want to use Albumentations for a specific task such as classification, segmentation, or object detection, refer to the set of articles that has an in-depth description of this task. We also have a list of examples on applying Albumentations for different use cases.
We have examples of using Albumentations along with PyTorch and TensorFlow.
Check the online demo of the library. With it, you can apply augmentations to different images and see the result. Also, we have a list of all available augmentations and their targets.
Pixel-level transforms will change just an input image and will leave any additional targets such as masks, bounding boxes, and keypoints unchanged. For volumetric data (volumes and 3D masks), these transforms are applied independently to each slice along the Z-axis (depth dimension), maintaining consistency across the volume. The list of pixel-level transforms:
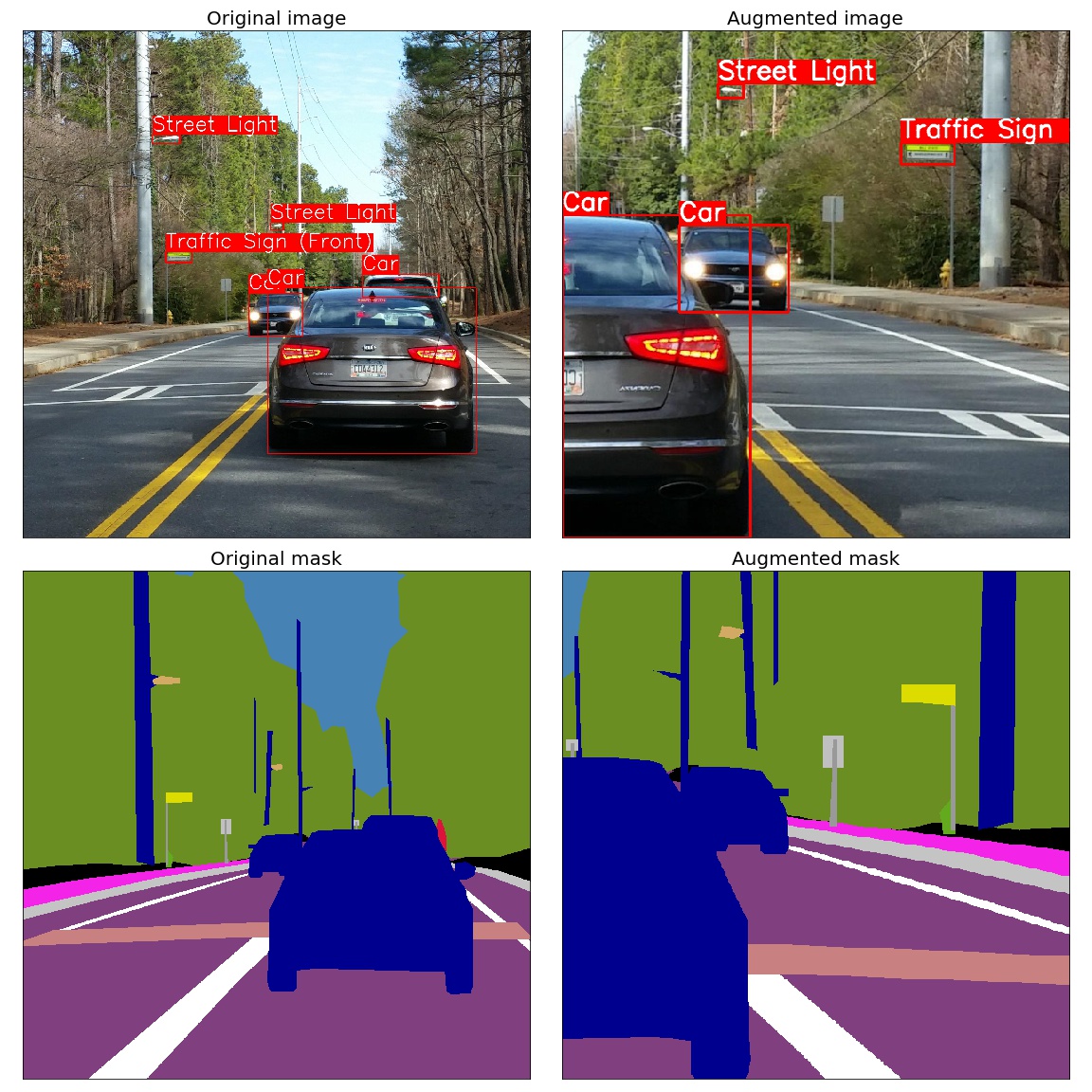
Spatial-level transforms will simultaneously change both an input image as well as additional targets such as masks, bounding boxes, and keypoints. For volumetric data (volumes and 3D masks), these transforms are applied independently to each slice along the Z-axis (depth dimension), maintaining consistency across the volume. The following table shows which additional targets are supported by each transform:
| Transform | Image | Mask | BBoxes | Keypoints | Volume | Mask3D |
|---|---|---|---|---|---|---|
| Affine | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| AtLeastOneBBoxRandomCrop | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| BBoxSafeRandomCrop | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| CenterCrop | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| CoarseDropout | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Crop | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| CropAndPad | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| CropNonEmptyMaskIfExists | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| D4 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
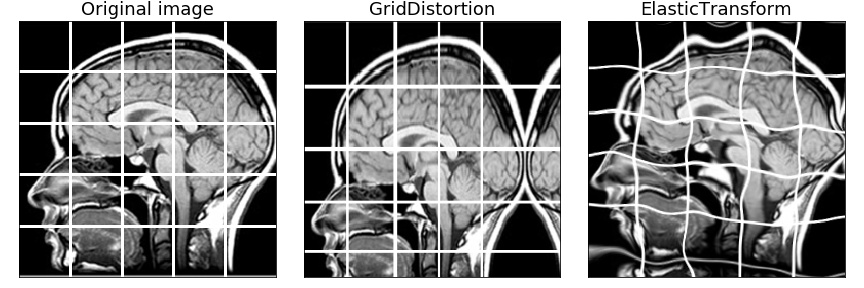
| ElasticTransform | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Erasing | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| FrequencyMasking | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| GridDistortion | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| GridDropout | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| GridElasticDeform | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| HorizontalFlip | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Lambda | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| LongestMaxSize | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| MaskDropout | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Morphological | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| NoOp | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| OpticalDistortion | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| OverlayElements | ✓ | ✓ | ||||
| Pad | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| PadIfNeeded | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Perspective | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| PiecewiseAffine | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| PixelDropout | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| RandomCrop | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| RandomCropFromBorders | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| RandomCropNearBBox | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| RandomGridShuffle | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| RandomResizedCrop | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| RandomRotate90 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| RandomScale | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| RandomSizedBBoxSafeCrop | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| RandomSizedCrop | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Resize | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Rotate | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| SafeRotate | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| ShiftScaleRotate | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| SmallestMaxSize | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| ThinPlateSpline | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| TimeMasking | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| TimeReverse | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Transpose | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| VerticalFlip | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| XYMasking | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
3D transforms operate on volumetric data and can modify both the input volume and associated 3D mask.
Where:
| Transform | Volume | Mask3D | Keypoints |
|---|---|---|---|
| CenterCrop3D | ✓ | ✓ | ✓ |
| CoarseDropout3D | ✓ | ✓ | ✓ |
| CubicSymmetry | ✓ | ✓ | ✓ |
| Pad3D | ✓ | ✓ | ✓ |
| PadIfNeeded3D | ✓ | ✓ | ✓ |
| RandomCrop3D | ✓ | ✓ | ✓ |

Number - is the number of uint8 RGB images processed per second on a single CPU core. Higher is better.
| Transform | albumentations 1.4.20 | augly 1.0.0 | imgaug 0.4.0 | kornia 0.7.3 | torchvision 0.20.0 |
|---|---|---|---|---|---|
| HorizontalFlip | 8618 ± 1233 | 4807 ± 818 | 6042 ± 788 | 390 ± 106 | 914 ± 67 |
| VerticalFlip | 22847 ± 2031 | 9153 ± 1291 | 10931 ± 1844 | 1212 ± 402 | 3198 ± 200 |
| Rotate | 1146 ± 79 | 1119 ± 41 | 1136 ± 218 | 143 ± 11 | 181 ± 11 |
| Affine | 682 ± 192 | - | 774 ± 97 | 147 ± 9 | 130 ± 12 |
| Equalize | 892 ± 61 | - | 581 ± 54 | 152 ± 19 | 479 ± 12 |
| RandomCrop80 | 47341 ± 20523 | 25272 ± 1822 | 11503 ± 441 | 1510 ± 230 | 32109 ± 1241 |
| ShiftRGB | 2349 ± 76 | - | 1582 ± 65 | - | - |
| Resize | 2316 ± 166 | 611 ± 78 | 1806 ± 63 | 232 ± 24 | 195 ± 4 |
| RandomGamma | 8675 ± 274 | - | 2318 ± 269 | 108 ± 13 | - |
| Grayscale | 3056 ± 47 | 2720 ± 932 | 1681 ± 156 | 289 ± 75 | 1838 ± 130 |
| RandomPerspective | 412 ± 38 | - | 554 ± 22 | 86 ± 11 | 96 ± 5 |
| GaussianBlur | 1728 ± 89 | 242 ± 4 | 1090 ± 65 | 176 ± 18 | 79 ± 3 |
| MedianBlur | 868 ± 60 | - | 813 ± 30 | 5 ± 0 | - |
| MotionBlur | 4047 ± 67 | - | 612 ± 18 | 73 ± 2 | - |
| Posterize | 9094 ± 301 | - | 2097 ± 68 | 430 ± 49 | 3196 ± 185 |
| JpegCompression | 918 ± 23 | 778 ± 5 | 459 ± 35 | 71 ± 3 | 625 ± 17 |
| GaussianNoise | 166 ± 12 | 67 ± 2 | 206 ± 11 | 75 ± 1 | - |
| Elastic | 201 ± 5 | - | 235 ± 20 | 1 ± 0 | 2 ± 0 |
| Clahe | 454 ± 22 | - | 335 ± 43 | 94 ± 9 | - |
| CoarseDropout | 13368 ± 744 | - | 671 ± 38 | 536 ± 87 | - |
| Blur | 5267 ± 543 | 246 ± 3 | 3807 ± 325 | - | - |
| ColorJitter | 628 ± 55 | 255 ± 13 | - | 55 ± 18 | 46 ± 2 |
| Brightness | 8956 ± 300 | 1163 ± 86 | - | 472 ± 101 | 429 ± 20 |
| Contrast | 8879 ± 1426 | 736 ± 79 | - | 425 ± 52 | 335 ± 35 |
| RandomResizedCrop | 2828 ± 186 | - | - | 287 ± 58 | 511 ± 10 |
| Normalize | 1196 ± 56 | - | - | 626 ± 40 | 519 ± 12 |
| PlankianJitter | 2204 ± 385 | - | - | 813 ± 211 | - |
To create a pull request to the repository, follow the documentation at CONTRIBUTING.md
If you find this library useful for your research, please consider citing Albumentations: Fast and Flexible Image Augmentations:
@Article{info11020125,
AUTHOR = {Buslaev, Alexander and Iglovikov, Vladimir I. and Khvedchenya, Eugene and Parinov, Alex and Druzhinin, Mikhail and Kalinin, Alexandr A.},
TITLE = {Albumentations: Fast and Flexible Image Augmentations},
JOURNAL = {Information},
VOLUME = {11},
YEAR = {2020},
NUMBER = {2},
ARTICLE-NUMBER = {125},
URL = {https://www.mdpi.com/2078-2489/11/2/125},
ISSN = {2078-2489},
DOI = {10.3390/info11020125}
}
FAQs
Fast, flexible, and advanced augmentation library for deep learning, computer vision, and medical imaging. Albumentations offers a wide range of transformations for both 2D (images, masks, bboxes, keypoints) and 3D (volumes, volumetric masks) data, with optimized performance and seamless integration into ML workflows.
We found that albumentations demonstrated a healthy version release cadence and project activity because the last version was released less than a year ago. It has 2 open source maintainers collaborating on the project.
Did you know?

Socket for GitHub automatically highlights issues in each pull request and monitors the health of all your open source dependencies. Discover the contents of your packages and block harmful activity before you install or update your dependencies.

Research
Security News
Socket researchers uncover a malicious npm package posing as a tool for detecting vulnerabilities in Etherium smart contracts.

Security News
Research
A supply chain attack on Rspack's npm packages injected cryptomining malware, potentially impacting thousands of developers.

Research
Security News
Socket researchers discovered a malware campaign on npm delivering the Skuld infostealer via typosquatted packages, exposing sensitive data.