Enrich_Omics: A Python wrapper for EnrichR and OpenTargets




Table of Contents
About
Python wrapper for EnrichR and OpenTargets API. Allows for visualisation of enriched pathways or diseases associated to a given target
EnrichR
- Choose from all EnrichR libraries (Transcription, Pathways, Drugs, etc). Default library is KEGG_pathways_2021
- Get table with enrichment results
- Plot enrichment results and export it
OpenTargets
Open Target is currently only supporting the search of a single target
Target endpoint
- Convert Entrez to Ensemble if needed (OpenTargets API accepts only Ensemble IDs)
- Get description of the function of the target
- Get diseases associated to a certain target
- Plot diseases associated to a certain target
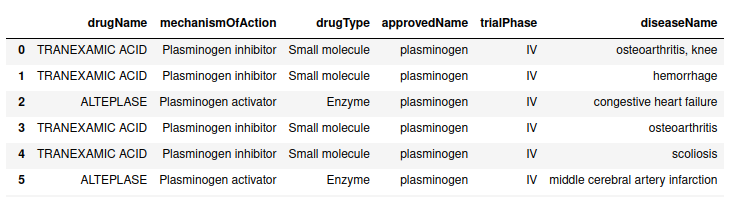
- Get table drugs associated to a certain target
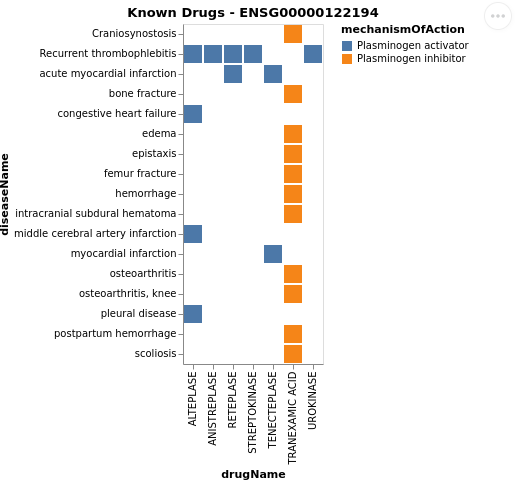
- Plot the drugs that work for a given target and the diseases associated to it
- Plot the drugs associated to a given target and the trial phase they are currently in
Plots
- Export plots in SVG and PNG
Installation
pip install enrich-omics
Tutorial
EnrichR API
import enrich_omics
from enrich_omics import EnrichR
from enrich_omics import OpenTargets
EnrichR.get_libraries()
gene_list = ['LMNA', 'MYH7', 'TNNT2', 'ACE2']
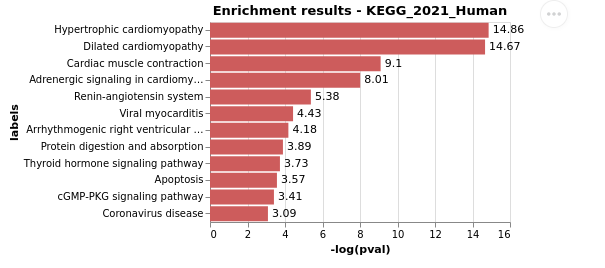
EnrichR.plot_enrichment(gene_list)
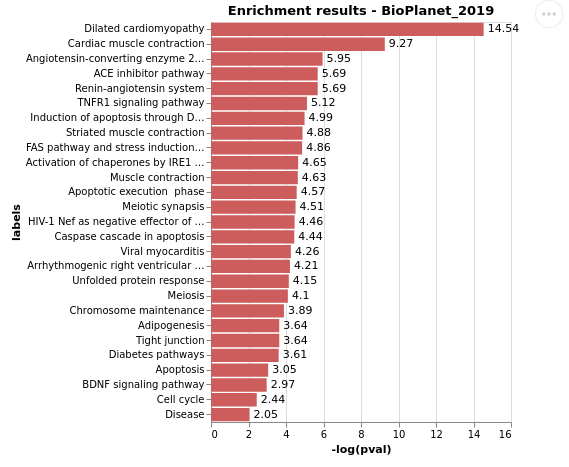
EnrichR.plot_enrichment(gene_list, library_name = 'BioPlanet_2019', height = 200, width = 300, max_hits= None)
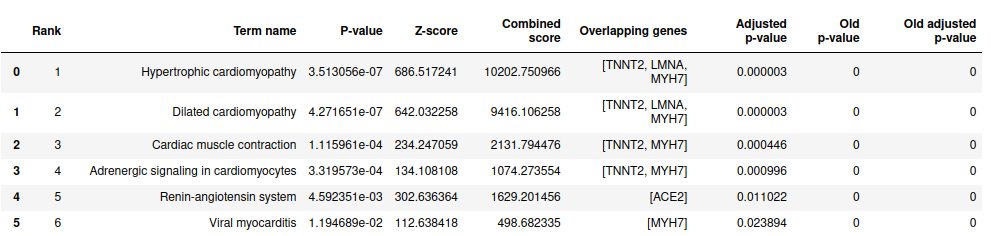
EnrichR.get_table_enrichment(['LMNA', 'MYH7', 'TNNT2', 'ACE2'], library_name='KEGG_2021_Human')
Open Targets API (Open Targets currently only supports single target enrichment. More information on the OpenTargets website)
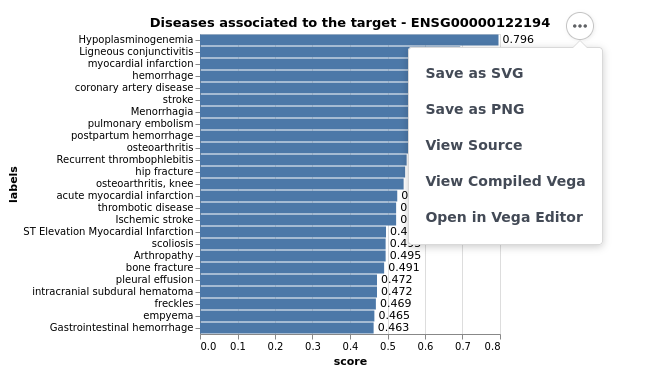
OpenTargets.plot_diseases(target_id = 'PLG', entrez = True)
OpenTargets.get_table_drugs(target_id = 'PLG', entrez = True)
OpenTargets.plot_drugs_disease(target_id = 'PLG', entrez = True)
Docs
Documentation for this package can be found here
Contributions always welcome!













