
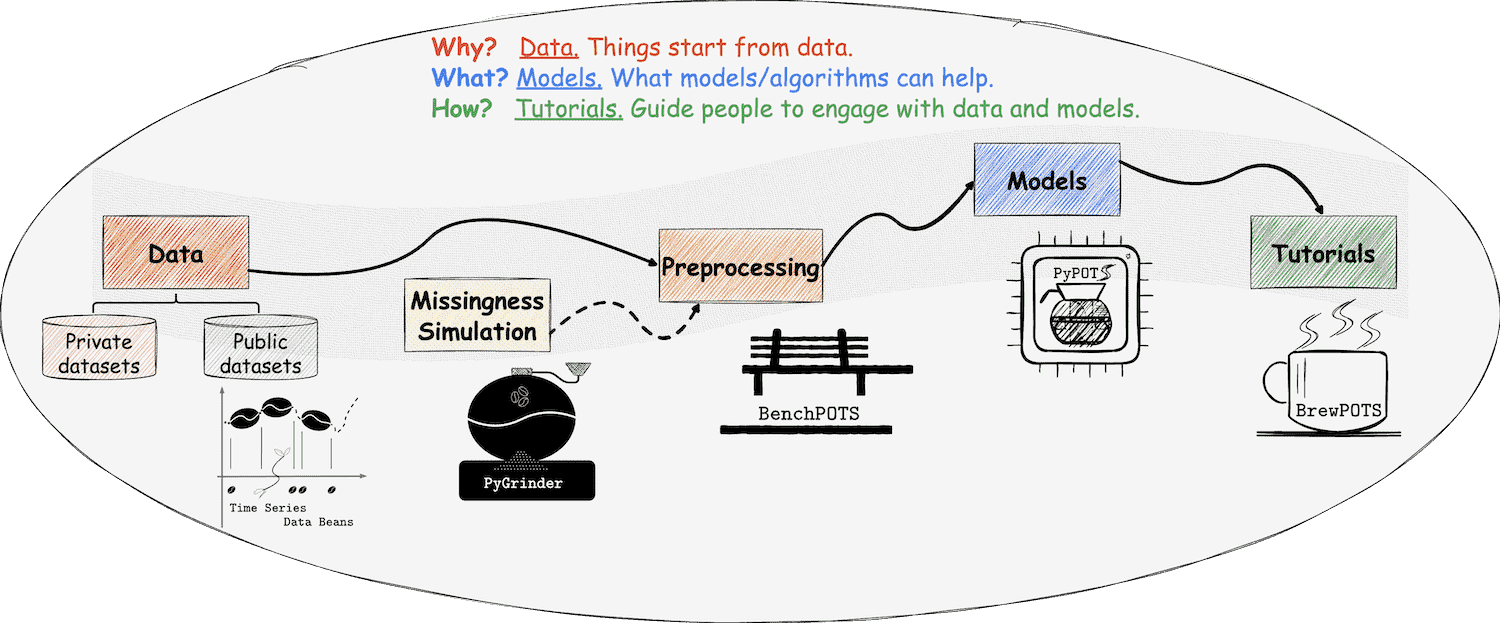
Welcome to PyGrinder
a Python toolkit for grinding data beans into the incomplete













 PyGrinder is a part of
PyPOTS
PyGrinder is a part of
PyPOTS  (a Python toolbox for data mining on
Partially-Observed Time Series), was called PyCorruptor and separated from PyPOTS for decoupling missingness-creating functionalities from
learning algorithms.
(a Python toolbox for data mining on
Partially-Observed Time Series), was called PyCorruptor and separated from PyPOTS for decoupling missingness-creating functionalities from
learning algorithms.
In data analysis and modeling, sometimes we may need to corrupt the original data to achieve our goal, for instance,
evaluating models' ability to reconstruct corrupted data or assessing the model's performance on only partially-observed
data. PyGrinder is such a tool to help you corrupt your data, which provides several patterns to create missing values
in the given data.
❖ Usage Examples
PyGrinder now is available on  ❗️
❗️
Install it with conda install pygrinder, you may need to specify the channel with option -c conda-forge
or install via PyPI:
pip install pygrinder
or install from source code:
pip install https://github.com/WenjieDu/PyGrinder/archive/main.zip
import numpy as np
from pygrinder import (
mcar,
mar_logistic,
mnar_x,
mnar_t,
mnar_nonuniform,
rdo,
seq_missing,
block_missing,
calc_missing_rate
)
ts_dataset = np.random.randn(128, 10, 36)
X_with_mcar_data = mcar(ts_dataset, p=0.1)
X_with_mar_data = mar_logistic(ts_dataset[:, 0, :], obs_rate=0.1, missing_rate=0.1)
X_with_mnar_x_data = mnar_x(ts_dataset, offset=0.1)
X_with_mnar_t_data = mnar_t(ts_dataset, cycle=20, pos=10, scale=3)
X_with_mnar_nonuniform_data = mnar_nonuniform(ts_dataset, p=0.5, increase_factor=0.5)
X_with_rdo_data = rdo(ts_dataset, p=0.1)
X_with_seq_missing_data = seq_missing(ts_dataset, p=0.1, seq_len=5)
X_with_block_missing_data = block_missing(ts_dataset, factor=0.1, block_width=3, block_len=3)
missing_rate = calc_missing_rate(X_with_mcar_data)
❖ Citing PyGrinder/PyPOTS
The paper introducing PyPOTS is available on arXiv,
A short version of it is accepted by the 9th SIGKDD international workshop on Mining and Learning from Time Series (MiLeTS'23)).
Additionally, PyPOTS has been included as a PyTorch Ecosystem project.
We are pursuing to publish it in prestigious academic venues, e.g. JMLR (track for
Machine Learning Open Source Software). If you use PyPOTS in your work,
please cite it as below and 🌟star this repository to make others notice this library. 🤗
There are scientific research projects using PyPOTS and referencing in their papers.
Here is an incomplete list of them.
@article{du2023pypots,
title={{PyPOTS: a Python toolbox for data mining on Partially-Observed Time Series}},
author={Wenjie Du},
journal={arXiv preprint arXiv:2305.18811},
year={2023},
}
or
Wenjie Du.
PyPOTS: a Python toolbox for data mining on Partially-Observed Time Series.
arXiv, abs/2305.18811, 2023.
🏠 Visits




















