PURPOSE
The most common gene cluster detection algorithms focus on canonical “core”
biosynthetic functions many gene clusters encode, while overlooking uncommon or
unknown cluster classes. These overlooked clusters are a potential source of
novel natural products and comprise an untold portion of overall gene cluster
repertoires. Unbiased, function-agnostic detection algorithms therefore provide
an opportunity to reveal novel classes of gene clusters and more broadly define
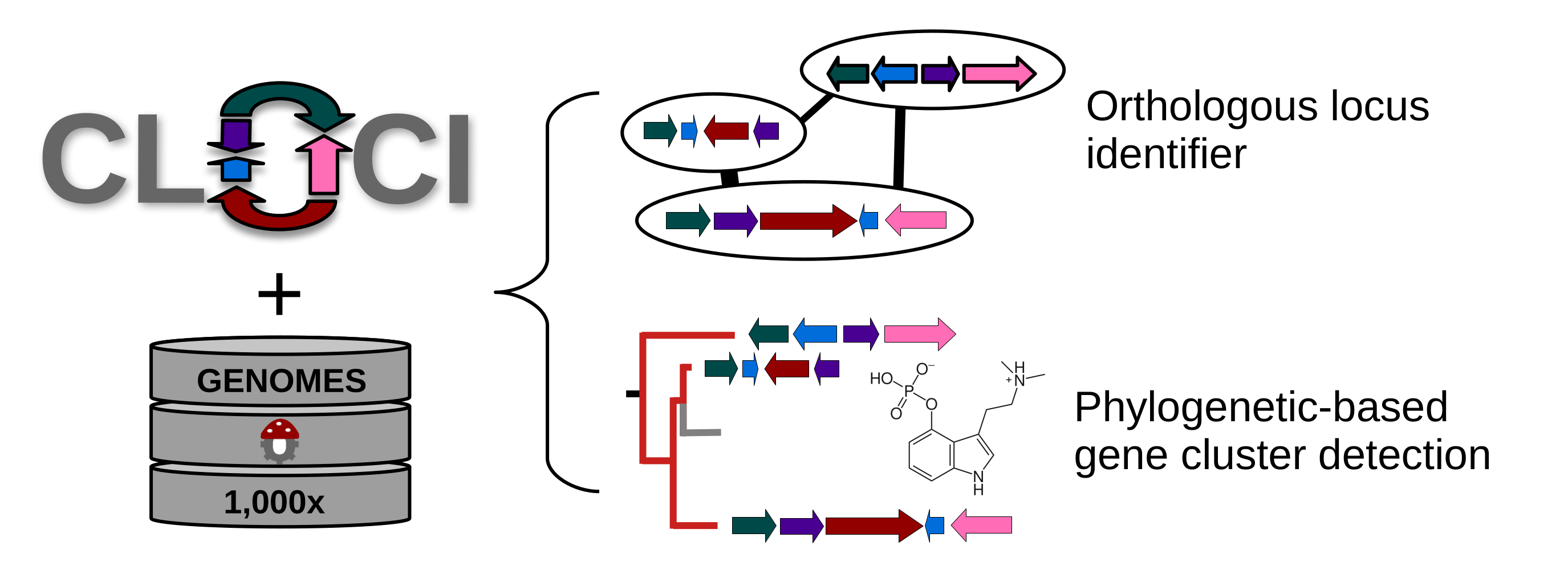
genome organization. CLOCI (Co-occurrence Locus and Orthologous Cluster
Identifier) is an algorithm that identifies gene clusters using multiple
proxies of selection for coordinated gene evolution. In the process, CLOCI
circumscribes loci into homologous locus groups, which is an extension of
orthogroups to the locus-level. Our approach generalizes gene cluster detection and gene cluster family circumscription, improves detection of multiple known functional classes, and unveils noncanonical gene clusters. CLOCI is suitable for genome-enabled specialized metabolite mining, and presents an easily tunable approach for delineating gene cluster families and homologous loci.
USAGE
Please see the wiki for installation
and usage instructions.
CITING
Zachary Konkel, Laura Kubatko, Jason C Slot, CLOCI: unveiling cryptic fungal
gene clusters with generalized detection, Nucleic Acids Research, 2024;, gkae625, https://doi.org/10.1093/nar/gkae625




