
Security News
vlt Launches "reproduce": A New Tool Challenging the Limits of Package Provenance
vlt's new "reproduce" tool verifies npm packages against their source code, outperforming traditional provenance adoption in the JavaScript ecosystem.
PlotHiC is used to visualize whole genome-wide contact heatmaps after genome scaffolding.
If you have any questions, please Open Issues or provide us with your comments via the email below.
Email: jzjlab@163.com
PlotHiC is utilised for the purpose of visualising genome-wide interaction heatmaps subsequent to de novo genome assembly. The software is compatible with both .hic and bed formats, and it is capable of adding chromosome names and custom visualization areas.
python = "^3.10"# pip install
pip install plothic
# create plothic enviorment and install plothic
conda env create -n plothic -c bioconda -c conda-forge plothic
# mamba env create -n plothic -c bioconda -c conda-forge plothic
If you want to see detailed usage and documentation of plothic, you can get it from wiki.
A simple example of PlotHiC use .hic(from Juicer/3D-DNA)and bed(from HiCPro)format is presented below.
genome.hicThis file is taken directly from 3d-dna, you need to select the final hic file (which has already been error adjusted and chromosome boundaries determined).
chr.tx (3 columns as follows, use "\t" as separator)Note: the length is in .hic file, not true base length (example as below).
# name length index
Chr1 24800000 5
Chr2 44380000 4
Chr3 63338000 3
Chr4 81187000 2
Chr5 97650000 1
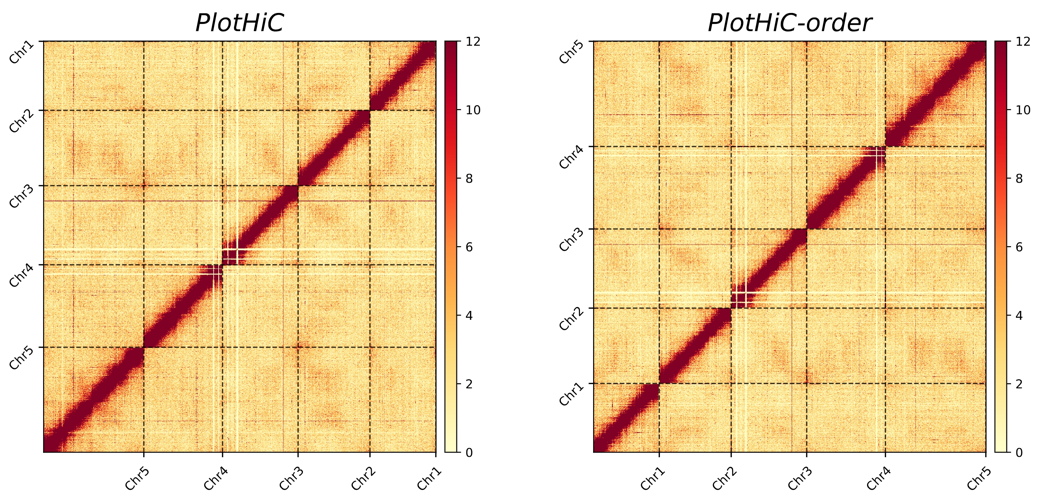
# Default order (left)
plothic -hic genome.hic -chr chr.txt -r 100000
# -hic > .hic file
# -chr > chromosome length (in .hic file)
# -r > resolution to visualization
# Custom order (right )
plothic -hic genome.hic -chr chr.txt -r 100000 --order
# --order > Sort by the order in chr.txt(index)
If the color performance is not to your liking, you can set parameters --bar-max to adjust it, which is very useful.
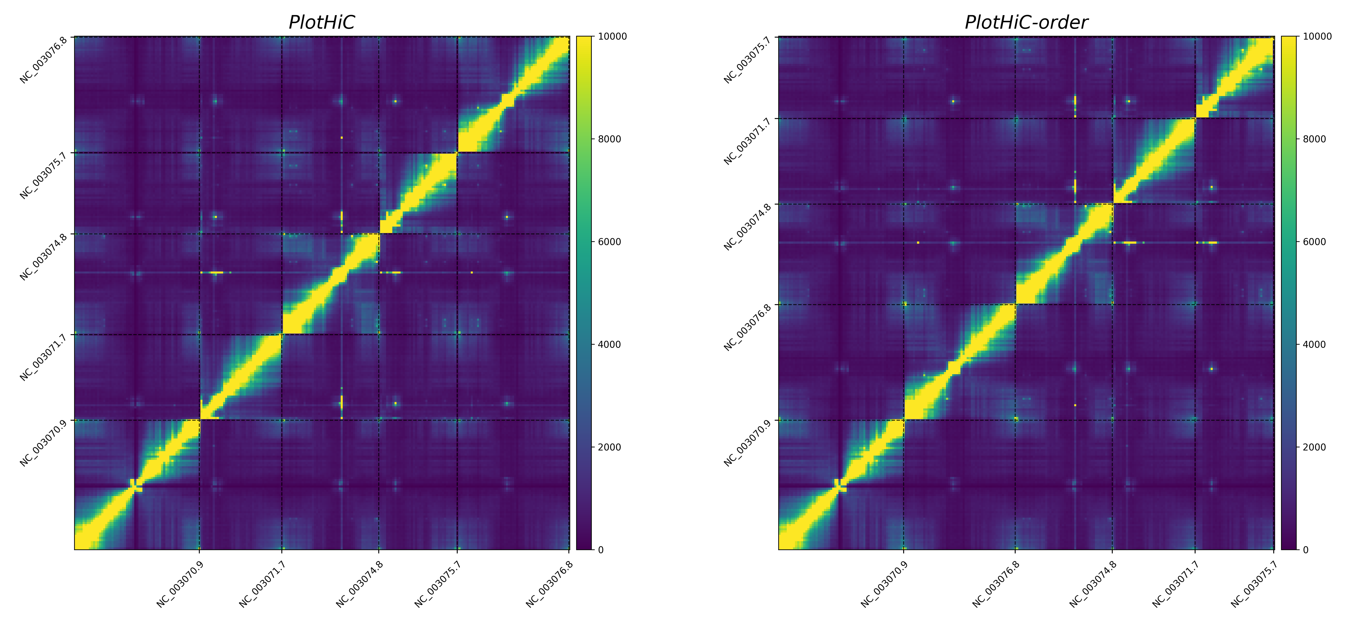
genome.matrixgenome_abs.bedBoth files are output from HiCPro. You can select a specific resolution based on your needs.
order.txt, the content and format as follows:# chr_name order
NC_003070.9 1
NC_003071.7 4
NC_003074.8 3
NC_003075.7 5
NC_003076.8 2
# Default order (left)
plothic -matrix sample_500000.matrix --abs-bed sample_500000_abs.bed -format png -cmap viridis --bar-max 10000 -g PlotHiC
# -matrix > matrix file
# --abs-bed > abs bed file
# -format > result format
# -cmap > viridis color
# --bar-max > max contact to show
# -g > Genome name to show top
# Custom order (right )
plothic -matrix sample_500000.matrix --abs-bed sample_500000_abs.bed -format png -cmap viridis --bar-max 10000 -g PlotHiC-order --abs-order order.txt
# --abs-order > Sort by the order in order.txt

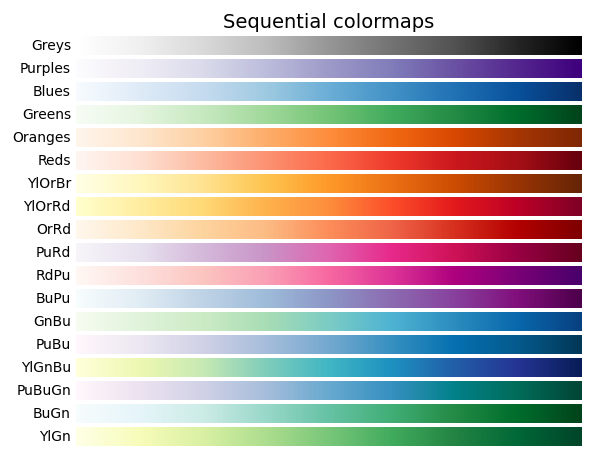
PlotHiC uses YlOrRd by default, you can choose more colors from Matplotlib.
If you used PlotHiC in your research, please cite us:
Zijie Jiang, Zhixiang Peng, Zhaoyuan Wei, Jiahe Sun, Yongjiang Luo, Lingzi Bie, Guoqing Zhang, Yi Wang, A deep learning-based method enables the automatic and accurate assembly of chromosome-level genomes, Nucleic Acids Research, 2024;, gkae789, https://doi.org/10.1093/nar/gkae789
FAQs
Plot Whole genome-wide Hi-C contact heatmap
We found that plothic demonstrated a healthy version release cadence and project activity because the last version was released less than a year ago. It has 1 open source maintainer collaborating on the project.
Did you know?

Socket for GitHub automatically highlights issues in each pull request and monitors the health of all your open source dependencies. Discover the contents of your packages and block harmful activity before you install or update your dependencies.

Security News
vlt's new "reproduce" tool verifies npm packages against their source code, outperforming traditional provenance adoption in the JavaScript ecosystem.

Research
Security News
Socket researchers uncovered a malicious PyPI package exploiting Deezer’s API to enable coordinated music piracy through API abuse and C2 server control.

Research
The Socket Research Team discovered a malicious npm package, '@ton-wallet/create', stealing cryptocurrency wallet keys from developers and users in the TON ecosystem.